Prolactin-induced protein is required for cell cycle progression in breast cancer
- PMID: 24862759
- PMCID: "V体育ios版" PMC4094838
- DOI: 10.1016/j.neo.2014.04.001
Prolactin-induced protein is required for cell cycle progression in breast cancer
Abstract
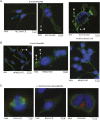
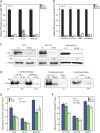
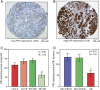
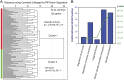
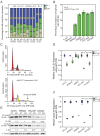
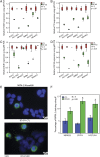
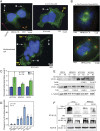
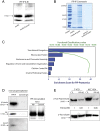
Prolactin-induced protein (PIP) is expressed in the majority of breast cancers and is used for the diagnostic evaluation of this disease as a characteristic biomarker; however, the molecular mechanisms of PIP function in breast cancer have remained largely unknown. In this study, we carried out a comprehensive investigation of PIP function using PIP silencing in a broad group of breast cancer cell lines, analysis of expression microarray data, proteomic analysis using mass spectrometry, and biomarker studies on breast tumors. We demonstrated that PIP is required for the progression through G1 phase, mitosis, and cytokinesis in luminal A, luminal B, and molecular apocrine breast cancer cells. In addition, PIP expression is associated with a transcriptional signature enriched with cell cycle genes and regulates key genes in this process including cyclin D1, cyclin B1, BUB1, and forkhead box M1 (FOXM1). It is notable that defects in mitotic transition and cytokinesis following PIP silencing are accompanied by an increase in aneuploidy of breast cancer cells. Importantly, we have identified novel PIP-binding partners in breast cancer and shown that PIP binds to β-tubulin and is necessary for microtubule polymerization. Furthermore, PIP interacts with actin-binding proteins including Arp2/3 and is needed for inside-out activation of integrin-β1 mediated through talin. This study suggests that PIP is required for cell cycle progression in breast cancer and provides a rationale for exploring PIP inhibition as a therapeutic approach in breast cancer that can potentially target microtubule polymerization. VSports手机版.
Copyright © 2014 Neoplasia Press, Inc. Published by Elsevier Inc. All rights reserved. V体育安卓版.
Figures


References
-
- Doane A.S., Danso M., Lal P., Donaton M., Zhang L., Hudis C., Gerald W.L. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. - PubMed
-
- Farmer P., Bonnefoi H., Becette V., Tubiana-Hulin M., Fumoleau P., Larsimont D., Macgrogan G., Bergh J., Cameron D., Goldstein D. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. - PubMed
Supplementary References
-
- Magklara A., Brown T.J., Diamandis E.P. Characterization of androgen receptor and nuclear receptor co-regulator expression in human breast cancer cell lines exhibiting differential regulation of kallikreins 2 and 3. Int J Cancer. 2002;100:507–514. - PubMed
-
- Heiser L.M., Sadanandam A., Kuo W.L., Benz S.C., Goldstein T.C., Ng S., Gibb W.J., Wang N.J., Ziyad S., Tong F. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–2729. - "V体育官网" PMC - PubMed
-
- Naderi A., Liu J. Inhibition of androgen receptor and Cdc25A phosphatase as a combination targeted therapy in molecular apocrine breast cancer. Cancer Lett. 2010;298:74–87. - PubMed
Publication types
- "VSports" Actions
- Actions (V体育平台登录)
MeSH terms
- Actions (V体育ios版)
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- VSports - Actions
- "V体育安卓版" Actions
- VSports app下载 - Actions
- "VSports在线直播" Actions
- Actions (VSports)
- "V体育安卓版" Actions
- Actions (VSports app下载)
"V体育平台登录" Substances
- V体育安卓版 - Actions
- VSports注册入口 - Actions
- "V体育ios版" Actions
Grants and funding (V体育官网)
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育官网)
Medical
Research Materials
Miscellaneous

