"VSports手机版" Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts
- PMID: 24732378
- PMCID: PMC4018493
- DOI: 10.1101/gad.239327.114 (VSports)
Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts
Abstract
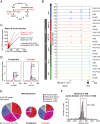
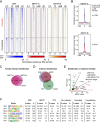
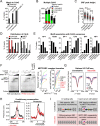
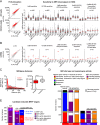
The transcription factor SRF (serum response factor) recruits two families of coactivators, the MRTFs (myocardin-related transcription factors) and the TCFs (ternary complex factors), to couple gene transcription to growth factor signaling. Here we investigated the role of the SRF network in the immediate transcriptional response of fibroblasts to serum stimulation. SRF recruited its cofactors in a gene-specific manner, and virtually all MRTF binding was directed by SRF. Much of SRF DNA binding was serum-inducible, reflecting a requirement for MRTF-SRF complex formation in nucleosome displacement. We identified 960 serum-responsive SRF target genes, which were mostly MRTF-controlled, as assessed by MRTF chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) and/or sensitivity to MRTF-linked signals. MRTF activation facilitates RNA polymerase II (Pol II) recruitment or promoter escape according to gene context. MRTF targets encode regulators of the cytoskeleton, transcription, and cell growth, underpinning the role of SRF in cytoskeletal dynamics and mechanosensing VSports手机版. Finally, we show that specific activation of either MRTFs or TCFs can reset the circadian clock. .
Keywords: MRTF; Rho; SRF; TCF; chromatin; signal transduction. V体育安卓版.
Figures


Comment in
-
Dual views of SRF: a genomic exposure.Genes Dev. 2014 May 1;28(9):926-8. doi: 10.1101/gad.242420.114. Genes Dev. 2014. PMID: 24788515 Free PMC article.
References
-
- Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ 2009. Complexity in transcription control at the activation domain-mediator interface. Sci Signal 2: ra20. - PMC (V体育安卓版) - PubMed
-
- Balsalobre A, Damiola F, Schibler U 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937 - PubMed
-
- Barker HE, Cox TR, Erler JT 2012. The rationale for targeting the LOX family in cancer. Nat Rev Cancer 12: 540–552 - PubMed
Publication types
MeSH terms
- Actions (V体育官网入口)
- VSports手机版 - Actions
- Actions (V体育2025版)
- Actions (VSports)
- "VSports app下载" Actions
- Actions (VSports手机版)
- Actions (VSports app下载)
Substances
- V体育官网 - Actions
- Actions (VSports注册入口)
Associated data
- "VSports在线直播" Actions
"V体育ios版" Grants and funding
VSports app下载 - LinkOut - more resources
Full Text Sources (VSports注册入口)
Other Literature Sources
Molecular Biology Databases
Miscellaneous
