Modulation of hypoxia-signaling pathways by extracellular linc-RoR
- PMID: 24463816
- PMCID: PMC3970562
- DOI: 10.1242/jcs.141069
Modulation of hypoxia-signaling pathways by extracellular linc-RoR
Abstract
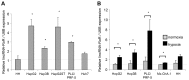
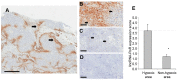
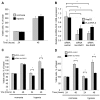
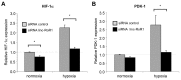
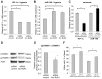
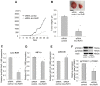
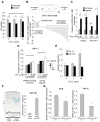
Resistance to adverse environmental conditions, such as hypoxia, contributes to the reduced efficacy of anticancer therapies and tumor progression. Although deregulated expression of many long noncoding RNA (lncRNA) occurs in human cancers, the contribution of such RNA to tumor responses to hypoxia are unknown. RNA expression profiling identified several hypoxia-responsive lncRNAs, including the long intergenic noncoding RNA, regulator of reprogramming (linc-RoR), which is also increased in expression in malignant liver cancer cells. Linc-RoR expression was increased in hypoxic regions within tumor cell xenografts in vivo. Tumor cell viability during hypoxia was reduced by small interfering RNA (siRNA) to linc-RoR. Compared with controls, siRNA to linc-RoR decreased phosphorylation of p70S6K1 (RPS6KB1), PDK1 and HIF-1α protein expression and increased expression of the linc-RoR target microRNA-145 (miR-145) VSports手机版. Linc-RoR was highly expressed in extracellular RNA released by hepatocellular cancer (HCC) cells during hypoxia. Incubation with extracellular vesicle preparations containing extracellular RNA increased linc-RoR, HIF-1α expression and cell survival in recipient cells. These studies show that linc-RoR is a hypoxia-responsive lncRNA that is functionally linked to hypoxia signaling in HCC through a miR-145-HIF-1α signaling module. Furthermore, this work identifies a mechanistic role for the extracellular transfer of linc-RoR in intercellular signaling to promote cell survival during hypoxic stress. .
Keywords: Cell stress; Exosome; Extracellular vesicle; Gene expression; Hypoxia; Liver cancer; linc-RoR; microRNA. V体育安卓版.
Figures

References
-
- Braconi C., Valeri N., Kogure T., Gasparini P., Huang N., Nuovo G. J., Terracciano L., Croce C. M., Patel T. (2011a). Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 108, 786–791 10.1073/pnas.1011098108 - DOI (V体育ios版) - PMC - PubMed
V体育平台登录 - Publication types
MeSH terms (V体育2025版)
- "V体育官网" Actions
- "V体育平台登录" Actions
- V体育官网 - Actions
- Actions (V体育安卓版)
- "VSports app下载" Actions
- V体育ios版 - Actions
- Actions (VSports注册入口)
- "V体育ios版" Actions
- Actions (VSports最新版本)
Substances
- Actions (V体育官网)
Grants and funding
LinkOut - more resources
V体育官网 - Full Text Sources
Other Literature Sources
Medical
VSports - Miscellaneous

