DNA polymerase η modulates replication fork progression and DNA damage responses in platinum-treated human cells
- PMID: 24253929
- PMCID: PMC6505966
- DOI: 10.1038/srep03277
"VSports最新版本" DNA polymerase η modulates replication fork progression and DNA damage responses in platinum-treated human cells
Abstract (V体育2025版)
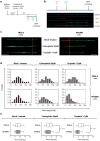
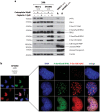
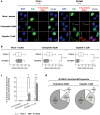
Human cells lacking DNA polymerase η (polη) are sensitive to platinum-based cancer chemotherapeutic agents. Using DNA combing to directly investigate the role of polη in bypass of platinum-induced DNA lesions in vivo, we demonstrate that nascent DNA strands are up to 39% shorter in human cells lacking polη than in cells expressing polη VSports手机版. This provides the first direct evidence that polη modulates replication fork progression in vivo following cisplatin and carboplatin treatment. Severe replication inhibition in individual platinum-treated polη-deficient cells correlates with enhanced phosphorylation of the RPA2 subunit of replication protein A on serines 4 and 8, as determined using EdU labelling and immunofluorescence, consistent with formation of DNA strand breaks at arrested forks in the absence of polη. Polη-mediated bypass of platinum-induced DNA lesions may therefore represent one mechanism by which cancer cells can tolerate platinum-based chemotherapy. .
Conflict of interest statement
The authors declare no competing financial interests.
VSports最新版本 - Figures

References
-
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7, 573–584 (2007). - PubMed (V体育ios版)
-
- Wernyj R. P. & Morin P. J. Molecular mechanisms of platinum resistance: still searching for the Achilles' heel. Drug Resist Update 7, 227–232 (2004). - V体育平台登录 - PubMed
-
- Wang D. & Lippard S. J. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 4, 307–320 (2005). - PubMed
-
- Chaney S. G., Campbell S. L., Bassett E. & Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol/Hematol 53, 3–11 (2005). - PubMed (VSports在线直播)
-
- Kartalou M. & Essigmann J. M. Recognition of cisplatin adducts by cellular proteins. Mutation Res/Fund Mol Mech Mutagen 478, 1–21 (2001). - "V体育2025版" PubMed
"VSports app下载" Publication types
MeSH terms
- Actions (VSports最新版本)
- Actions (VSports app下载)
- Actions (VSports最新版本)
- VSports - Actions
- VSports注册入口 - Actions
- Actions (VSports)
- V体育官网 - Actions
- V体育官网入口 - Actions
- "VSports app下载" Actions
- "VSports注册入口" Actions
- Actions (VSports app下载)
Substances
- "V体育2025版" Actions
- "V体育ios版" Actions
LinkOut - more resources
V体育安卓版 - Full Text Sources
VSports手机版 - Other Literature Sources
Molecular Biology Databases

