Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells
- PMID: 23031797
- PMCID: VSports手机版 - PMC3534407
- DOI: 10.1186/1471-2407-12-445
Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells
Abstract
Background: Breast cancer and its metastatic progression is mainly directed by epithelial to mesenchymal transition (EMT), a phenomenon supported by specific transcription factors and miRNAs VSports手机版. .
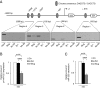
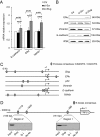
Methods: In order to investigate a possible correlation between Slug transcription factor and miR-221, we performed Slug gene silencing in MDA-MB-231 breast cancer cells and evaluated the expression of genes involved in supporting the breast cancer phenotype, using qRT-PCR and Western blot analysis. Chromatin immunoprecipitation and wound healing assays were employed to determine a functional link between these two molecules V体育安卓版. .
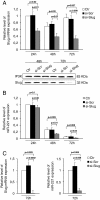
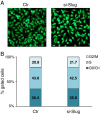
Results: We showed that Slug silencing significantly decreased the level of miR-221 and vimentin, reactivated Estrogen Receptor α and increased E-cadherin and TRPS1 expression V体育ios版. We demonstrated that miR-221 is a Slug target gene, and identified a specific region of miR-221 promoter that is transcriptionally active and binds the transcription factor Slug "in vivo". In addition, we showed that in Slug-silenced cells, wich retained residual miR-221 (about 38%), cell migration was strongly inhibited. Cell migration was inhibited, but to a less degree, following complete knockdown of miR-221 expression by transfection with antagomiR-221. .
Conclusions: We report for the first time evidence of a correlation between Slug transcription factor and miR-221 in breast cancer cells VSports最新版本. These studies suggest that miR-221 expression is, in part, dependent on Slug in breast cancer cells, and that Slug plays a more important role than miR-221 in cell migration and invasion. .
Figures

References
-
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. - "V体育官网" DOI - PubMed
-
- Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5(2):101–106. doi: 10.1186/bcr578. - V体育官网 - DOI - PMC - PubMed
-
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. - V体育2025版 - DOI - PubMed
Publication types (V体育官网入口)
- "VSports在线直播" Actions
MeSH terms
- VSports app下载 - Actions
- VSports最新版本 - Actions
- "V体育官网" Actions
- VSports最新版本 - Actions
- VSports最新版本 - Actions
- Actions (VSports最新版本)
- V体育ios版 - Actions
VSports最新版本 - Substances
- VSports注册入口 - Actions
- VSports最新版本 - Actions
"VSports" LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

