The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends (V体育安卓版)
- PMID: 22960743
- PMCID: PMC3640768
- DOI: 10.1038/nature11355
The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends (VSports手机版)
Abstract (V体育2025版)
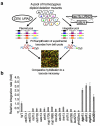
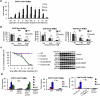
Chromosomal double-strand breaks (DSBs) are resected by 5' nucleases to form 3' single-stranded DNA substrates for binding by homologous recombination and DNA damage checkpoint proteins. Two redundant pathways of extensive resection have been described both in cells and in vitro, one relying on Exo1 exonuclease and the other on Sgs1 helicase and Dna2 nuclease VSports手机版. However, it remains unknown how resection proceeds within the context of chromatin, where histones and histone-bound proteins represent barriers for resection enzymes. Here we identify the yeast nucleosome-remodelling enzyme Fun30 as a factor promoting DSB end resection. Fun30 is the major nucleosome remodeller promoting extensive Exo1- and Sgs1-dependent resection of DSBs. The RSC and INO80 chromatin-remodelling complexes and Fun30 have redundant roles in resection adjacent to DSB ends. ATPase and helicase domains of Fun30, which are needed for nucleosome remodelling, are also required for resection. Fun30 is robustly recruited to DNA breaks and spreads along the DSB coincident with resection. Fun30 becomes less important for resection in the absence of the histone-bound Rad9 checkpoint adaptor protein known to block 5' strand processing and in the absence of either histone H3 K79 methylation or γ-H2A, which mediate recruitment of Rad9 (refs 9, 10). Together these data suggest that Fun30 helps to overcome the inhibitory effect of Rad9 on DNA resection. .
Figures


"VSports注册入口" References
-
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. - "VSports手机版" PMC - PubMed
Publication types
- Actions (V体育平台登录)
MeSH terms
- Actions (VSports最新版本)
- "VSports app下载" Actions
- "VSports最新版本" Actions
- "V体育2025版" Actions
- "V体育官网" Actions
- V体育安卓版 - Actions
- "VSports在线直播" Actions
- VSports在线直播 - Actions
- "VSports手机版" Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
- "V体育平台登录" Actions
- "V体育官网" Actions
- Actions (V体育平台登录)
"V体育2025版" Substances
- Actions (VSports注册入口)
- "VSports在线直播" Actions
- Actions (V体育官网)
- VSports - Actions
- Actions (VSports最新版本)
- "VSports" Actions
"V体育ios版" Associated data
Grants and funding
"VSports" LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

