IDH mutation impairs histone demethylation and results in a block to cell differentiation
- PMID: 22343901
- PMCID: PMC3478770
- DOI: 10.1038/nature10860
IDH mutation impairs histone demethylation and results in a block to cell differentiation
Abstract
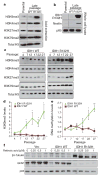
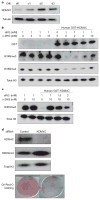
Recurrent mutations in isocitrate dehydrogenase 1 (IDH1) and IDH2 have been identified in gliomas, acute myeloid leukaemias (AML) and chondrosarcomas, and share a novel enzymatic property of producing 2-hydroxyglutarate (2HG) from α-ketoglutarate VSports手机版. Here we report that 2HG-producing IDH mutants can prevent the histone demethylation that is required for lineage-specific progenitor cells to differentiate into terminally differentiated cells. In tumour samples from glioma patients, IDH mutations were associated with a distinct gene expression profile enriched for genes expressed in neural progenitor cells, and this was associated with increased histone methylation. To test whether the ability of IDH mutants to promote histone methylation contributes to a block in cell differentiation in non-transformed cells, we tested the effect of neomorphic IDH mutants on adipocyte differentiation in vitro. Introduction of either mutant IDH or cell-permeable 2HG was associated with repression of the inducible expression of lineage-specific differentiation genes and a block to differentiation. This correlated with a significant increase in repressive histone methylation marks without observable changes in promoter DNA methylation. Gliomas were found to have elevated levels of similar histone repressive marks. Stable transfection of a 2HG-producing mutant IDH into immortalized astrocytes resulted in progressive accumulation of histone methylation. Of the marks examined, increased H3K9 methylation reproducibly preceded a rise in DNA methylation as cells were passaged in culture. Furthermore, we found that the 2HG-inhibitable H3K9 demethylase KDM4C was induced during adipocyte differentiation, and that RNA-interference suppression of KDM4C was sufficient to block differentiation. Together these data demonstrate that 2HG can inhibit histone demethylation and that inhibition of histone demethylation can be sufficient to block the differentiation of non-transformed cells. .
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www. nature. com/nature V体育安卓版.
"V体育官网入口" Figures


V体育2025版 - Comment in
-
VSports最新版本 - Metabolism: unmasking an oncometabolite.Nat Rev Cancer. 2012 Mar 1;12(4):229. doi: 10.1038/nrc3248. Nat Rev Cancer. 2012. PMID: 22378191 No abstract available.
"VSports手机版" References
-
- Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. - "V体育安卓版" PMC - PubMed
-
- Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. - PubMed
-
- Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. - "VSports在线直播" PMC - PubMed
Publication types
- "VSports手机版" Actions
MeSH terms
- "V体育官网" Actions
- VSports在线直播 - Actions
- Actions (VSports最新版本)
- Actions (V体育ios版)
- V体育平台登录 - Actions
- V体育平台登录 - Actions
- V体育安卓版 - Actions
- "VSports在线直播" Actions
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- "VSports手机版" Actions
- Actions (VSports)
- "V体育安卓版" Actions
- Actions (V体育2025版)
- Actions (VSports最新版本)
- "V体育2025版" Actions
Substances
- "V体育平台登录" Actions
- Actions (VSports在线直播)
- "V体育2025版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
V体育安卓版 - Molecular Biology Databases
Miscellaneous

