Molecular profiling of patient-derived breast cancer xenografts
- PMID: 22247967
- PMCID: PMC3496128
- DOI: 10.1186/bcr3095
Molecular profiling of patient-derived breast cancer xenografts
Abstract
Introduction: Identification of new therapeutic agents for breast cancer (BC) requires preclinical models that reproduce the molecular characteristics of their respective clinical tumors. In this work, we analyzed the genomic and gene expression profiles of human BC xenografts and the corresponding patient tumors. VSports手机版.
Methods: Eighteen BC xenografts were obtained by grafting tumor fragments from patients into Swiss nude mice. Molecular characterization of patient tumors and xenografts was performed by DNA copy number analysis and gene expression analysis using Affymetrix Microarrays. V体育安卓版.
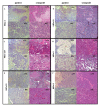
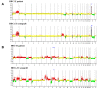
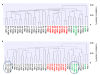
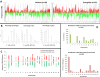
Results: Comparison analysis showed that 14/18 pairs of tumors shared more than 56% of copy number alterations (CNA). Unsupervised hierarchical clustering analysis showed that 16/18 pairs segregated together, confirming the similarity between tumor pairs. Analysis of recurrent CNA changes between patient tumors and xenografts showed losses in 176 chromosomal regions and gains in 202 chromosomal regions. Gene expression profile analysis showed that less than 5% of genes had recurrent variations between patient tumors and their respective xenografts; these genes largely corresponded to human stromal compartment genes. Finally, analysis of different passages of the same tumor showed that sequential mouse-to-mouse tumor grafts did not affect genomic rearrangements or gene expression profiles, suggesting genetic stability of these models over time. V体育ios版.
Conclusions: This panel of human BC xenografts maintains the overall genomic and gene expression profile of the corresponding patient tumors and remains stable throughout sequential in vivo generations. The observed genomic profile and gene expression differences appear to be due to the loss of human stromal genes. These xenografts, therefore, represent a validated model for preclinical investigation of new therapeutic agents. VSports最新版本.
V体育官网入口 - Figures


References
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI (VSports app下载) - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - VSports - DOI - PMC - PubMed
-
- Garber K. From human to mouse and back: 'tumorgraft' models surge in popularity. J Natl Cancer Inst. 2009;101:6–8. - PubMed
Publication types
- VSports注册入口 - Actions
MeSH terms
- Actions (V体育2025版)
- "V体育2025版" Actions
- "V体育ios版" Actions
- Actions (VSports注册入口)
- VSports注册入口 - Actions
- "VSports注册入口" Actions
- Actions (VSports)
Substances (V体育平台登录)
- "V体育官网" Actions
LinkOut - more resources
Full Text Sources
VSports - Medical
"V体育2025版" Research Materials
Miscellaneous

