"VSports最新版本" The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression
- PMID: 21967847
- PMCID: PMC3273810
- DOI: 10.1093/nar/gkr818
"V体育ios版" The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression
Abstract
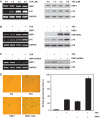
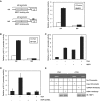
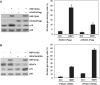
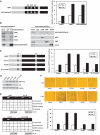
HBP1 is a sequence-specific DNA-binding transcription factor with many important biological roles. It activates or represses the expression of some specific genes during cell growth and differentiation. Previous studies have exhibited that HBP1 binds to p16(INK4A) promoter and activates p16(INK4A) expression. We found that trichostatin A (TSA), an inhibitor of HDAC (histone deacetylase), induces p16(INK4A) expression in an HBP1-dependent manner. This result was drawn from a transactivation experiment by measuring relative luciferase activities of p16(INK4A) promoter with HBP1-binding site in comparison with that of the wild-type p16(INK4A) promoter by transient cotransfection with HBP1 into HEK293T cells and 2BS cells. HBP1 acetylation after TSA treatment was confirmed by immunoprecipitation assay. Our data showed that HBP1 interacted with histone acetyltransferase p300 and CREB-binding protein (CBP) and also recruited p300/CBP to p16(INK4A) promoter. HBP1 was acetylated by p300/CBP in two regions: repression domain (K297/305/307) and P domain (K171/419). Acetylation of Repression domain was not required for HBP1 transactivation on p16(INK4A). However, luciferase assay and western blotting results indicate that acetylation of P domain, especially K419 acetylation is essential for HBP1 transactivation on p16(INK4A) VSports手机版. As assayed by SA-beta-gal staining, the acetylation of HBP1 at K419 enhanced HBP1-induced premature senescence in 2BS cells. In addition, HDAC4 repressed HBP1-induced premature senescence through permanently deacetylating HBP1. We conclude that our data suggest that HBP1 acetylation at K419 plays an important role in HBP1-induced p16(INK4A) expression. .
"VSports手机版" Figures





References
-
- Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, et al. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. - PubMed
-
- Lavender P, Vandel L, Bannister AJ, Kouzarides T. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene. 1997;14:2721–2728. - PubMed
Publication types
- "VSports app下载" Actions
MeSH terms
- "VSports app下载" Actions
- V体育官网 - Actions
- VSports注册入口 - Actions
- VSports - Actions
- Actions (VSports注册入口)
- Actions (V体育安卓版)
- V体育ios版 - Actions
- VSports在线直播 - Actions
- Actions (VSports在线直播)
- "V体育2025版" Actions
- Actions (VSports)
- "VSports在线直播" Actions
Substances
- "V体育官网入口" Actions
- V体育平台登录 - Actions
- "V体育官网" Actions
- Actions (VSports最新版本)
- "V体育ios版" Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
LinkOut - more resources
"V体育官网入口" Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous (V体育2025版)

