Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein (VSports app下载)
- PMID: 21880555
- PMCID: PMC3185188 (VSports最新版本)
- DOI: 10.1016/j.dnarep.2011.08.002
"V体育官网" Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein
VSports最新版本 - Abstract
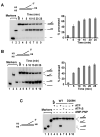
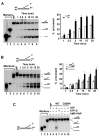
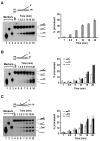
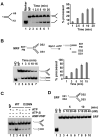
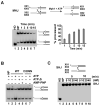
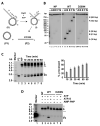
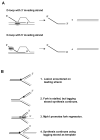
The budding yeast Mph1 protein, the putative ortholog of human FANCM, possesses a 3' to 5' DNA helicase activity and is capable of disrupting the D-loop structure to suppress chromosome arm crossovers in mitotic homologous recombination. Similar to FANCM, genetic studies have implicated Mph1 in DNA replication fork repair VSports手机版. Consistent with this genetic finding, we show here that Mph1 is able to mediate replication fork reversal, and to process the Holliday junction via DNA branch migration. Moreover, Mph1 unwinds 3' and 5' DNA Flap structures that bear key features of the D-loop. These biochemical results not only provide validation for a role of Mph1 in the repair of damaged replication forks, but they also offer mechanistic insights as to its ability to efficiently disrupt the D-loop intermediate. .
Copyright © 2011 Elsevier B. V. All rights reserved V体育安卓版. .
Figures

References
-
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. - PMC (V体育安卓版) - PubMed
-
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. - PubMed
"V体育官网" Publication types
"V体育ios版" MeSH terms
- Actions (VSports注册入口)
- Actions (V体育官网)
- VSports注册入口 - Actions
- V体育2025版 - Actions
- "VSports手机版" Actions
- "V体育官网入口" Actions
Substances
- V体育安卓版 - Actions
- Actions (V体育官网入口)
- "VSports在线直播" Actions
"V体育平台登录" Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

