Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1
- PMID: 21789191
- PMCID: "V体育官网入口" PMC3138737
- DOI: 10.1371/journal.pone.0021889
Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1
"VSports app下载" Abstract
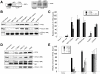
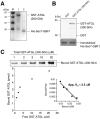
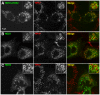
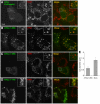
The Arf1 exchange factor GBF1 (Golgi Brefeldin A resistance factor 1) and its effector COPI are required for delivery of ATGL (adipose triglyceride lipase) to lipid droplets (LDs). Using yeast two hybrid, co-immunoprecipitation in mammalian cells and direct protein binding approaches, we report here that GBF1 and ATGL interact directly and in cells, through multiple contact sites on each protein. The C-terminal region of ATGL interacts with N-terminal domains of GBF1, including the catalytic Sec7 domain, but not with full-length GBF1 or its entire N-terminus. The N-terminal lipase domain of ATGL (called the patatin domain) interacts with two C-terminal domains of GBF1, HDS (Homology downstream of Sec7) 1 and HDS2 VSports手机版. These two domains of GBF1 localize to lipid droplets when expressed alone in cells, but not to the Golgi, unlike the full-length GBF1 protein, which localizes to both. We suggest that interaction of GBF1 with ATGL may be involved in the membrane trafficking pathway mediated by GBF1, Arf1 and COPI that contributes to the localization of ATGL to lipid droplets. .
Conflict of interest statement
Figures





"VSports" References
-
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
-
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. - "VSports手机版" PubMed
-
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. - PubMed
-
- Deng Y, Golinelli-Cohen MP, Smirnova E, Jackson CL. A COPI coat subunit interacts directly with an early-Golgi localized Arf exchange factor. EMBO Rep. 2009;10:58–64. - VSports app下载 - PMC - PubMed
"V体育平台登录" Publication types
- Actions (V体育ios版)
MeSH terms
- Actions (V体育ios版)
- Actions (V体育官网入口)
- Actions (V体育2025版)
- Actions (V体育官网入口)
- "V体育2025版" Actions
- VSports最新版本 - Actions
- V体育官网 - Actions
- "V体育安卓版" Actions
Substances (VSports注册入口)
- "VSports最新版本" Actions
- V体育安卓版 - Actions
- Actions (VSports app下载)
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

