Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication
- PMID: 21784862
- PMCID: PMC3173079 (V体育平台登录)
- DOI: 10.1074/jbc.M111.258038
Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication
Erratum in
- J Biol Chem. 2011 Nov 11;286(45):39674
VSports手机版 - Abstract
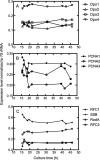
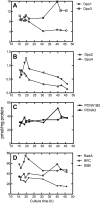
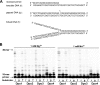
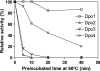
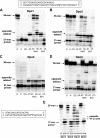
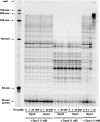
The hyperthermophilic crenarchaeon Sulfolobus solfataricus P2 encodes three B-family DNA polymerase genes, B1 (Dpo1), B2 (Dpo2), and B3 (Dpo3), and one Y-family DNA polymerase gene, Dpo4, which are related to eukaryotic counterparts. Both mRNAs and proteins of all four DNA polymerases were constitutively expressed in all growth phases. Dpo2 and Dpo3 possessed very low DNA polymerase and 3' to 5' exonuclease activities in vitro. Steady-state kinetic efficiencies (k(cat)/K(m)) for correct nucleotide insertion by Dpo2 and Dpo3 were several orders of magnitude less than Dpo1 and Dpo4 VSports手机版. Both the accessory proteins proliferating cell nuclear antigen and the clamp loader replication factor C facilitated DNA synthesis with Dpo3, as with Dpo1 and Dpo4, but very weakly with Dpo2. DNA synthesis by Dpo2 and Dpo3 was remarkably decreased by single-stranded binding protein, in contrast to Dpo1 and Dpo4. DNA synthesis in the presence of proliferating cell nuclear antigen, replication factor C, and single-stranded binding protein was most processive with Dpo1, whereas DNA lesion bypass was most effective with Dpo4. Both Dpo2 and Dpo3, but not Dpo1, bypassed hypoxanthine and 8-oxoguanine. Dpo2 and Dpo3 bypassed uracil and cis-syn cyclobutane thymine dimer, respectively. High concentrations of Dpo2 or Dpo3 did not attenuate DNA synthesis by Dpo1 or Dpo4. We conclude that Dpo2 and Dpo3 are much less functional and more thermolabile than Dpo1 and Dpo4 in vitro but have bypass activities across hypoxanthine, 8-oxoguanine, and either uracil or cis-syn cyclobutane thymine dimer, suggesting their catalytically limited roles in translesion DNA synthesis past deaminated, oxidized base lesions and/or UV-induced damage. .
VSports - Figures








References
-
- Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., W.H. Freeman, New York
-
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, 2nd Ed., American Society for Microbiology Press, Washington, D. C
-
- Rothwell P. J., Waksman G. (2005) Adv. Protein Chem. 71, 401–440 - PubMed
-
- Hubscher U., Maga G., Spadari S. (2002) Annu. Rev. Biochem. 71, 133–163 - VSports注册入口 - PubMed
-
- Rivera M. C., Lake J. A. (2004) Nature 431, 152–155 - PubMed
Publication types
MeSH terms
- V体育官网入口 - Actions
- "V体育安卓版" Actions
- Actions (VSports在线直播)
- Actions (VSports手机版)
- Actions (VSports app下载)
- V体育官网入口 - Actions
- "V体育安卓版" Actions
- V体育官网入口 - Actions
- "V体育2025版" Actions
- Actions (V体育平台登录)
Substances (VSports最新版本)
- VSports注册入口 - Actions
- VSports - Actions
- "VSports最新版本" Actions
- VSports最新版本 - Actions
- Actions (V体育2025版)
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

