Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth
- PMID: 21219636
- PMCID: "V体育官网" PMC3109565
- DOI: 10.1186/bcr2803
Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth (VSports最新版本)
V体育ios版 - Abstract
Introduction: MicroRNAs (miRNAs) are a class of small non-coding RNAs (20 to 24 nucleotides) that post-transcriptionally modulate gene expression. A key oncomir in carcinogenesis is miR-21, which is consistently up-regulated in a wide range of cancers. However, few functional studies are available for miR-21, and few targets have been identified VSports手机版. In this study, we explored the role of miR-21 in human breast cancer cells and tissues, and searched for miR-21 targets. .
Methods: We used in vitro and in vivo assays to explore the role of miR-21 in the malignant progression of human breast cancer, using miR-21 knockdown. Using LNA silencing combined to microarray technology and target prediction, we screened for potential targets of miR-21 and validated direct targets by using luciferase reporter assay and Western blot V体育安卓版. Two candidate target genes (EIF4A2 and ANKRD46) were selected for analysis of correlation with clinicopathological characteristics and prognosis using immunohistochemistry on cancer tissue microrrays. .
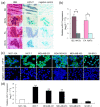
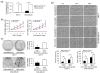
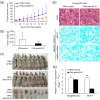
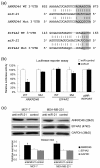
Results: Anti-miR-21 inhibited growth and migration of MCF-7 and MDA-MB-231 cells in vitro, and tumor growth in nude mice. Knockdown of miR-21 significantly increased the expression of ANKRD46 at both mRNA and protein levels. Luciferase assays using a reporter carrying a putative target site in the 3' untranslated region of ANKRD46 revealed that miR-21 directly targeted ANKRD46. miR-21 and EIF4A2 protein were inversely expressed in breast cancers (rs = -0. 283, P = 0. 005, Spearman's correlation analysis) V体育ios版. .
Conclusions: Knockdown of miR-21 in MCF-7 and MDA-MB-231 cells inhibits in vitro and in vivo growth as well as in vitro migration VSports最新版本. ANKRD46 is newly identified as a direct target of miR-21 in BC. These results suggest that inhibitory strategies against miR-21 using peptide nucleic acids (PNAs)-antimiR-21 may provide potential therapeutic applications in breast cancer treatment. .
Figures

References
-
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. - "V体育平台登录" DOI - PubMed
-
- Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. - V体育2025版 - DOI - PubMed
Publication types
MeSH terms
- Actions (VSports注册入口)
- "V体育ios版" Actions
- VSports手机版 - Actions
- V体育官网 - Actions
- V体育2025版 - Actions
- Actions (VSports在线直播)
- Actions (V体育ios版)
- Actions (VSports最新版本)
- Actions (V体育2025版)
- V体育2025版 - Actions
- VSports在线直播 - Actions
- "VSports注册入口" Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- Actions (VSports)
- VSports - Actions
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
V体育安卓版 - Miscellaneous

