"V体育平台登录" A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress
- PMID: 21187895
- PMCID: "V体育安卓版" PMC3002981
- DOI: 10.1371/journal.pgen.1001247
A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress (VSports最新版本)
Erratum in
- PLoS Genet. 2011 Feb;7(2). doi: 10.1371/annotation/6549d0b1-efde-4aa4-9cda-1cef43f66b30
Abstract
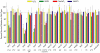
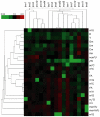
Decades of study have revealed more than 100 ribonucleoside structures incorporated as post-transcriptional modifications mainly in tRNA and rRNA, yet the larger functional dynamics of this conserved system are unclear. To this end, we developed a highly precise mass spectrometric method to quantify tRNA modifications in Saccharomyces cerevisiae. Our approach revealed several novel biosynthetic pathways for RNA modifications and led to the discovery of signature changes in the spectrum of tRNA modifications in the damage response to mechanistically different toxicants. This is illustrated with the RNA modifications Cm, m(5)C, and m(2) (2)G, which increase following hydrogen peroxide exposure but decrease or are unaffected by exposure to methylmethane sulfonate, arsenite, and hypochlorite. Cytotoxic hypersensitivity to hydrogen peroxide is conferred by loss of enzymes catalyzing the formation of Cm, m(5)C, and m(2) (2)G, which demonstrates that tRNA modifications are critical features of the cellular stress response. The results of our study support a general model of dynamic control of tRNA modifications in cellular response pathways and add to the growing repertoire of mechanisms controlling translational responses in cells VSports手机版. .
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
Better living through biochemistry.Nat Methods. 2011 Feb;8(2):108-9. doi: 10.1038/nmeth0211-108b. Nat Methods. 2011. PMID: 21355118 No abstract available.
References
-
- Alberts B. Extended version. New York: Garland Science; 2008. Molecular biology of the cell, 5th edition.
-
- Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. - PubMed (VSports app下载)
-
- Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391–16395. - PubMed
"VSports" Publication types
- "V体育官网入口" Actions
- "V体育平台登录" Actions
MeSH terms
- "VSports注册入口" Actions
- "V体育官网" Actions
- "V体育ios版" Actions
- Actions (V体育2025版)
Substances
- Actions (VSports手机版)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

