Minimising immunohistochemical false negative ER classification using a complementary 23 gene expression signature of ER status
- PMID: 21152022
- PMCID: PMC2995741
- DOI: VSports - 10.1371/journal.pone.0015031
Minimising immunohistochemical false negative ER classification using a complementary 23 gene expression signature of ER status (VSports app下载)
Abstract
Background: Expression of the oestrogen receptor (ER) in breast cancer predicts benefit from endocrine therapy VSports手机版. Minimising the frequency of false negative ER status classification is essential to identify all patients with ER positive breast cancers who should be offered endocrine therapies in order to improve clinical outcome. In routine oncological practice ER status is determined by semi-quantitative methods such as immunohistochemistry (IHC) or other immunoassays in which the ER expression level is compared to an empirical threshold. The clinical relevance of gene expression-based ER subtypes as compared to IHC-based determination has not been systematically evaluated. Here we attempt to reduce the frequency of false negative ER status classification using two gene expression approaches and compare these methods to IHC based ER status in terms of predictive and prognostic concordance with clinical outcome. .
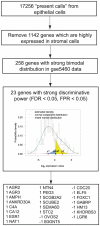
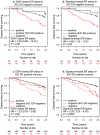
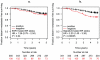
Methodology/principal findings: Firstly, ER status was discriminated by fitting the bimodal expression of ESR1 to a mixed Gaussian model. The discriminative power of ESR1 suggested bimodal expression as an efficient way to stratify breast cancer; therefore we identified a set of genes whose expression was both strongly bimodal, mimicking ESR expression status, and highly expressed in breast epithelial cell lines, to derive a 23-gene ER expression signature-based classifier. We assessed our classifiers in seven published breast cancer cohorts by comparing the gene expression-based ER status to IHC-based ER status as a predictor of clinical outcome in both untreated and tamoxifen treated cohorts V体育安卓版. In untreated breast cancer cohorts, the 23 gene signature-based ER status provided significantly improved prognostic power compared to IHC-based ER status (P = 0. 006). In tamoxifen-treated cohorts, the 23 gene ER expression signature predicted clinical outcome (HR = 2. 20, P = 0. 00035). These complementary ER signature-based strategies estimated that between 15. 1% and 21. 8% patients of IHC-based negative ER status would be classified with ER positive breast cancer. .
Conclusion/significance: Expression-based ER status classification may complement IHC to minimise false negative ER status classification and optimise patient stratification for endocrine therapies. V体育ios版.
"VSports最新版本" Conflict of interest statement
Figures

References
-
- Barnes DM MR, Beex LV, Thorpe SM, Leake RE. Increased use of immunohistochemistry for oestrogen receptor measurement in mammary carcinoma: the need for quality assurance. European Journal of Cancer. 1998;34 - PubMed
-
- Weigelt B, Mackay A, A'Hern R, Natrajan R, Tan DS, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol - PubMed
-
- Arpino G, Weiss H, Lee AV, Schiff R, De Placido S, et al. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J Natl Cancer Inst. 2005;97:1254–1261. - PubMed
-
- Gown AM. Current issues in ER and HER2 testing by IHC in breast cancer. Mod Pathol. 2008;21(Suppl 2):S8–S15. - PubMed (V体育2025版)
-
- Gong Y, Yan K, Lin F, Anderson K, Sotiriou C, et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: a gene-expression profiling study. Lancet Oncol. 2007;8:203–211. - PubMed
Publication types
- "VSports最新版本" Actions
MeSH terms (V体育2025版)
- "V体育2025版" Actions
- "V体育安卓版" Actions
- VSports注册入口 - Actions
- V体育ios版 - Actions
- V体育官网 - Actions
- VSports注册入口 - Actions
- V体育安卓版 - Actions
- "V体育平台登录" Actions
- "V体育官网入口" Actions
Substances
"V体育平台登录" Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases (VSports手机版)
Miscellaneous

