Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7
- PMID: 21070963
- PMCID: PMC3021128
- DOI: 10.1016/j.molcel.2010.10.017 (V体育ios版)
Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7
Abstract
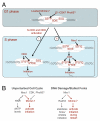
Activation of the eukaryotic replicative DNA helicase, the Mcm2-7 complex, requires phosphorylation by Cdc7/Dbf4 (Dbf4-dependent kinase or DDK), which, in turn, depends on prior phosphorylation of Mcm2-7 by an unknown kinase (or kinases). We identified DDK phosphorylation sites on Mcm4 and Mcm6 and found that phosphorylation of either subunit suffices for cell proliferation. Importantly, prior phosphorylation of either S/T-P or S/T-Q motifs on these subunits is required for DDK phosphorylation of Mcm2-7 and for normal S phase passage. Phosphomimetic mutations of DDK target sites bypass both DDK function and mutation of the priming phosphorylation sites VSports手机版. Mrc1 facilitates Mec1 phosphorylation of the S/T-Q motifs of chromatin-bound Mcm2-7 during S phase to activate replication. Genetic interactions between priming site mutations and MRC1 or TOF1 deletion support a role for these modifications in replication fork stability. These findings identify regulatory mechanisms that modulate origin firing and replication fork assembly during cell cycle progression. .
Copyright © 2010 Elsevier Inc V体育安卓版. All rights reserved. .
Figures


References
-
- Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem. 2009;284:28823–28831. - "V体育官网入口" PMC - PubMed
-
- Charych DH, Coyne M, Yabannavar A, Narberes J, Chow S, Wallroth M, Shafer C, Walter AO. Inhibition of Cdc7/Dbf4 kinase activity affects specific phosphorylation sites on MCM2 in cancer cells. J Cell Biochem. 2008;104:1075–1086. - V体育官网入口 - PubMed
-
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103:11521–11526. - VSports在线直播 - PMC - PubMed
-
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. - PubMed
Publication types
MeSH terms
- Actions (VSports)
- Actions (VSports在线直播)
- V体育ios版 - Actions
- "VSports app下载" Actions
- VSports app下载 - Actions
- Actions (V体育官网)
- Actions (VSports手机版)
- V体育2025版 - Actions
- "VSports" Actions
- "VSports注册入口" Actions
Substances
- "V体育平台登录" Actions
- Actions (VSports最新版本)
- Actions (V体育安卓版)
- "V体育官网入口" Actions
- "VSports手机版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
V体育ios版 - Miscellaneous

