Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism
- PMID: 20979384
- PMCID: PMC3010197
- DOI: V体育平台登录 - 10.1021/bi1012518
Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism
Abstract
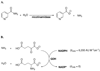
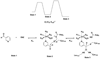
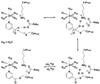
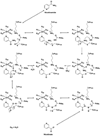
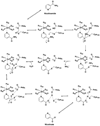
Nicotinamidases are metabolic enzymes that hydrolyze nicotinamide to nicotinic acid. These enzymes are widely distributed across biology, with examples found encoded in the genomes of Mycobacteria, Archaea, Eubacteria, Protozoa, yeast, and invertebrates, but there are none found in mammals. Although recent structural work has improved our understanding of these enzymes, their catalytic mechanism is still not well understood. Recent data show that nicotinamidases are required for the growth and virulence of several pathogenic microbes. The enzymes of Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans regulate life span in their respective organisms, consistent with proposed roles in the regulation of NAD(+) metabolism and organismal aging. In this work, the steady state kinetic parameters of nicotinamidase enzymes from C. elegans, Sa. cerevisiae, Streptococcus pneumoniae (a pathogen responsible for human pneumonia), Borrelia burgdorferi (the pathogen that causes Lyme disease), and Plasmodium falciparum (responsible for most human malaria) are reported. Nicotinamidases are generally efficient catalysts with steady state k(cat) values typically exceeding 1 s(-1). The K(m) values for nicotinamide are low and in the range of 2 -110 μM VSports手机版. Nicotinaldehyde was determined to be a potent competitive inhibitor of these enzymes, binding in the low micromolar to low nanomolar range for all nicotinamidases tested. A variety of nicotinaldehyde derivatives were synthesized and evaluated as inhibitors in kinetic assays. Inhibitions are consistent with reaction of the universally conserved catalytic Cys on each enzyme with the aldehyde carbonyl carbon to form a thiohemiacetal complex that is stabilized by a conserved oxyanion hole. The S. pneumoniae nicotinamidase can catalyze exchange of (18)O into the carboxy oxygens of nicotinic acid with H(2)(18)O. The collected data, along with kinetic analysis of several mutants, allowed us to propose a catalytic mechanism that explains nicotinamidase and nicotinic acid (18)O exchange chemistry for the S. pneumoniae enzyme involving key catalytic residues, a catalytic transition metal ion, and the intermediacy of a thioester intermediate. .
Figures (VSports app下载)








References
-
- Gerdes SY, Scholle MD, D'Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. - "V体育官网入口" PMC - PubMed
-
- Zhang H, Deng JY, Bi LJ, Zhou YF, Zhang ZP, Zhang CG, Zhang Y, Zhang XE. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. Febs J. 2008;275:753–762. - VSports app下载 - PubMed
-
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. - PubMed
-
- Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–224. - PubMed
Publication types
- VSports在线直播 - Actions
- VSports app下载 - Actions
- Actions (V体育ios版)
MeSH terms
- "V体育官网" Actions
- Actions (VSports app下载)
- VSports手机版 - Actions
- "V体育官网" Actions
- Actions (V体育平台登录)
- Actions (V体育2025版)
- Actions (VSports注册入口)
"VSports最新版本" Substances
- VSports - Actions
- V体育安卓版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
V体育官网 - Chemical Information
Molecular Biology Databases
Miscellaneous (V体育2025版)

