"V体育安卓版" Comprehensive comparative analysis of strand-specific RNA sequencing methods
- PMID: 20711195
- PMCID: PMC3005310
- DOI: V体育安卓版 - 10.1038/nmeth.1491
Comprehensive comparative analysis of strand-specific RNA sequencing methods
"VSports在线直播" Abstract
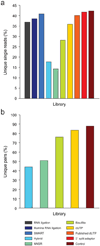
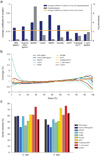
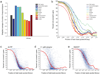
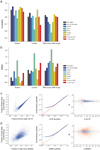
Strand-specific, massively parallel cDNA sequencing (RNA-seq) is a powerful tool for transcript discovery, genome annotation and expression profiling. There are multiple published methods for strand-specific RNA-seq, but no consensus exists as to how to choose between them. Here we developed a comprehensive computational pipeline to compare library quality metrics from any RNA-seq method. Using the well-annotated Saccharomyces cerevisiae transcriptome as a benchmark, we compared seven library-construction protocols, including both published and our own methods. We found marked differences in strand specificity, library complexity, evenness and continuity of coverage, agreement with known annotations and accuracy for expression profiling. Weighing each method's performance and ease, we identified the dUTP second-strand marking and the Illumina RNA ligation methods as the leading protocols, with the former benefitting from the current availability of paired-end sequencing VSports手机版. Our analysis provides a comprehensive benchmark, and our computational pipeline is applicable for assessment of future protocols in other organisms. .
Figures


References
-
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. - "V体育官网" PMC - PubMed
-
- Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. - PubMed
-
- Yassour M, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:3264–3269. - VSports app下载 - PMC - PubMed
-
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. - "V体育ios版" PMC - PubMed
Publication types
V体育ios版 - MeSH terms
- V体育安卓版 - Actions
- Actions (V体育官网)
- VSports手机版 - Actions
- Actions (V体育平台登录)
- VSports - Actions
Substances
- V体育官网入口 - Actions
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
"VSports最新版本" Other Literature Sources
V体育官网入口 - Molecular Biology Databases

