Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies
- PMID: 20671025
- PMCID: PMC2995073
- DOI: 10.1093/nar/gkq655
"V体育官网" Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies
Abstract
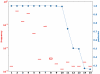
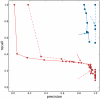
Next-generation sequencing technologies can be used to analyse genetically heterogeneous samples at unprecedented detail. The high coverage achievable with these methods enables the detection of many low-frequency variants. However, sequencing errors complicate the analysis of mixed populations and result in inflated estimates of genetic diversity. We developed a probabilistic Bayesian approach to minimize the effect of errors on the detection of minority variants. We applied it to pyrosequencing data obtained from a 1. 5-kb-fragment of the HIV-1 gag/pol gene in two control and two clinical samples. The effect of PCR amplification was analysed. Error correction resulted in a two- and five-fold decrease of the pyrosequencing base substitution rate, from 0. 05% to 0. 03% and from 0. 25% to 0. 05% in the non-PCR and PCR-amplified samples, respectively. We were able to detect viral clones as rare as 0. 1% with perfect sequence reconstruction. Probabilistic haplotype inference outperforms the counting-based calling method in both precision and recall. Genetic diversity observed within and between two clinical samples resulted in various patterns of phenotypic drug resistance and suggests a close epidemiological link. We conclude that pyrosequencing can be used to investigate genetically diverse samples with high accuracy if technical errors are properly treated VSports手机版. .
Figures



References
-
- Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. - PubMed (VSports最新版本)
-
- Miller W, Drautz DI, Ratan A, Pusey B, Qi J, Lesk AM, Tomsho LP, Packard MD, Zhao F, Sher A, et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456:387–390. - PubMed
-
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. - PubMed
Publication types (VSports)
- V体育2025版 - Actions
MeSH terms
- VSports在线直播 - Actions
- Actions (VSports注册入口)
- Actions (VSports注册入口)
- "V体育官网" Actions
- "VSports" Actions
Substances
LinkOut - more resources (V体育官网)
V体育平台登录 - Full Text Sources
Other Literature Sources

