Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase
- PMID: 20194642
- PMCID: PMC2835936
- DOI: 10.1083/jcb.200905059
Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase
Abstract
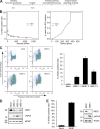
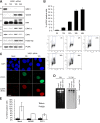
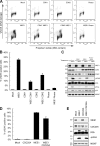
Maintenance of genome integrity is of critical importance to cells VSports手机版. To identify key regulators of genomic integrity, we screened a human cell line with a kinome small interfering RNA library. WEE1, a major regulator of mitotic entry, and CHK1 were among the genes identified. Both kinases are important negative regulators of CDK1 and -2. Strikingly, WEE1 depletion rapidly induced DNA damage in S phase in newly replicated DNA, which was accompanied by a marked increase in single-stranded DNA. This DNA damage is dependent on CDK1 and -2 as well as the replication proteins MCM2 and CDT1 but not CDC25A. Conversely, DNA damage after CHK1 inhibition is highly dependent on CDC25A. Furthermore, the inferior proliferation of CHK1-depleted cells is improved substantially by codepletion of CDC25A. We conclude that the mitotic kinase WEE1 and CHK1 jointly maintain balanced cellular control of Cdk activity during normal DNA replication, which is crucial to prevent the generation of harmful DNA lesions during replication. .
Figures (V体育官网)

VSports - Comment in
-
High-throughput siRNA screens using γH2AX as marker uncover key regulators of genome integrity in mammalian cells.Cell Cycle. 2010 Jun 15;9(12):2257-8. doi: 10.4161/cc.9.12.12017. Epub 2010 Jun 15. Cell Cycle. 2010. PMID: 20699633 No abstract available.
References (V体育ios版)
-
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637 10.1038/nature05268 - DOI - PubMed
-
- Chen Y., Poon R.Y. 2008. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front. Biosci. 13:5016–5029 - PubMed
V体育ios版 - Publication types
- V体育ios版 - Actions
MeSH terms
- Actions (V体育平台登录)
- Actions (VSports手机版)
- "V体育2025版" Actions
- "V体育ios版" Actions
- V体育官网 - Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- VSports app下载 - Actions
- Actions (VSports app下载)
- V体育安卓版 - Actions
- "VSports最新版本" Actions
Substances
- "V体育平台登录" Actions
- "VSports手机版" Actions
- VSports最新版本 - Actions
- "VSports" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

