The YTH domain is a novel RNA binding domain
- PMID: 20167602
- PMCID: PMC2863249
- DOI: "V体育官网入口" 10.1074/jbc.M110.104711
The YTH domain is a novel RNA binding domain
Abstract (V体育平台登录)
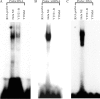
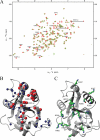
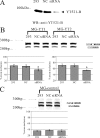
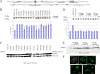
The YTH (YT521-B homology) domain was identified by sequence comparison and is found in 174 different proteins expressed in eukaryotes. It is characterized by 14 invariant residues within an alpha-helix/beta-sheet structure. Here we show that the YTH domain is a novel RNA binding domain that binds to a short, degenerated, single-stranded RNA sequence motif. The presence of the binding motif in alternative exons is necessary for YT521-B to directly influence splice site selection in vivo. Array analyses demonstrate that YT521-B predominantly regulates vertebrate-specific exons. An NMR titration experiment identified the binding surface for single-stranded RNA on the YTH domain. Structural analyses indicate that the YTH domain is related to the pseudouridine synthase and archaeosine transglycosylase (PUA) domain. Our data show that the YTH domain conveys RNA binding ability to a new class of proteins that are found in all eukaryotic organisms VSports手机版. .
"VSports" Figures



References
-
- Chen Y., Varani G. (2005) FEBS J. 272, 2088–2097 - PubMed
-
- Maris C., Dominguez C., Allain F. H. (2005) FEBS J. 272, 2118–2131 - PubMed
-
- Pérez-Arellano I., Gallego J., Cervera J. (2007) FEBS J. 274, 4972–4984 - PubMed
-
- Lie Y. S., Macdonald P. M. (1999) Development 126, 1129–1138 - PubMed (VSports app下载)
Publication types (V体育2025版)
MeSH terms
- "VSports在线直播" Actions
- "VSports app下载" Actions
- V体育平台登录 - Actions
- "V体育官网入口" Actions
- Actions (V体育平台登录)
- V体育ios版 - Actions
- Actions (VSports手机版)
- VSports最新版本 - Actions
Substances
- Actions (VSports在线直播)
- "VSports在线直播" Actions
VSports注册入口 - LinkOut - more resources
Full Text Sources
Molecular Biology Databases

