Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis
- PMID: 20110263
- PMCID: PMC2875002
- DOI: 10.1093/nar/gkq012
VSports最新版本 - Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis
Abstract
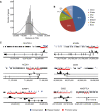
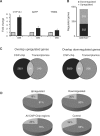
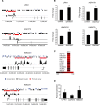
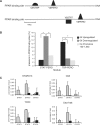
The transcription factor peroxisome proliferator-activated receptor alpha (PPARalpha) is an important regulator of hepatic lipid metabolism VSports手机版. While PPARalpha is known to activate transcription of numerous genes, no comprehensive picture of PPARalpha binding to endogenous genes has yet been reported. To fill this gap, we performed Chromatin immunoprecipitation (ChIP)-chip in combination with transcriptional profiling on HepG2 human hepatoma cells treated with the PPARalpha agonist GW7647. We found that GW7647 increased PPARalpha binding to 4220 binding regions. GW7647-induced binding regions showed a bias around the transcription start site and most contained a predicted PPAR binding motif. Several genes known to be regulated by PPARalpha, such as ACOX1, SULT2A1, ACADL, CD36, IGFBP1 and G0S2, showed GW7647-induced PPARalpha binding to their promoter. A GW7647-induced PPARalpha-binding region was also assigned to SREBP-targets HMGCS1, HMGCR, FDFT1, SC4MOL, and LPIN1, expression of which was induced by GW7647, suggesting cross-talk between PPARalpha and SREBP signaling. Our data furthermore demonstrate interaction between PPARalpha and STAT transcription factors in PPARalpha-mediated transcriptional repression, and suggest interaction between PPARalpha and TBP, and PPARalpha and C/EBPalpha in PPARalpha-mediated transcriptional activation. Overall, our analysis leads to important new insights into the mechanisms and impact of transcriptional regulation by PPARalpha in human liver and highlight the importance of cross-talk with other transcription factors. .
Figures

References
-
- Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. - PubMed
-
- Mandard S, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol. Life Sci. 2004;61:393–416. - "VSports注册入口" PMC - PubMed
-
- Zandbergen F, Plutzky J. PPARalpha in atherosclerosis and inflammation. Biochim. Biophys. Acta. 2007;1771:972–982. - PMC (V体育官网入口) - PubMed
-
- Michalik L, Wahli W. PPARs mediate lipid signaling in inflammation and cancer. PPAR Res. 2008;2008:134059. - "VSports" PMC - PubMed
Publication types
- VSports app下载 - Actions
MeSH terms
- Actions (V体育官网)
- V体育平台登录 - Actions
- Actions (V体育官网)
- "VSports在线直播" Actions
- Actions (V体育官网入口)
Substances
- "V体育官网入口" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

