"V体育2025版" Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults
- PMID: 19508560
- PMCID: PMC2784037
- DOI: "V体育ios版" 10.1111/j.1462-2920.2009.01900.x
Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults
"V体育ios版" Abstract
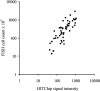
In this paper we present the in silico assessment of the diversity of variable regions of the small subunit ribosomal RNA (SSU rRNA) gene based on an ecosystem-specific curated database, describe a probe design procedure based on two hypervariable regions with minimal redundancy and test the potential of such probe design strategy for the design of a flexible microarray platform. This resulted in the development and application of a phylogenetic microarray for studying the human gastrointestinal microbiota--referred as the human intestinal tract chip (HITChip). Over 4800 dedicated tiling oligonucleotide probes were designed based on two hypervariable regions of the SSU rRNA gene of 1140 unique microbial phylotypes (< 98% identity) following analysis of over 16,000 human intestinal SSU rRNA sequences. These HITChip probes were hybridized to a diverse set of human intestinal samples and SSU rRNA clones to validate its fingerprinting and quantification potential. Excellent reproducibility (median Pearson's correlation of 0. 99) was obtained following hybridization with T7 polymerase transcripts generated in vitro from SSU rRNA gene amplicons. A linear dose-response was observed with artificial mixtures of 40 different representative amplicons with relative abundances as low as 0. 1% of total microbiota. Analysis of three consecutively collected faecal samples from ten individuals (five young and five elderly adults) revealed temporal dynamics and confirmed that the adult intestinal microbiota is an individual-specific and relatively stable ecosystem. Further analysis of the stable part allowed for the identification of a universal microbiota core at the approximate genus level (90% sequence similarity). This core consists of members of Actinobacteria, Bacteroidetes and Firmicutes. Used as a phylogenetic fingerprinting tool with the possibility for relative quantification, the HITChip has the potential to bridge the gaps in our knowledge in the quantitative and qualitative description of the human gastrointestinal microbiota composition VSports手机版. .
Figures (VSports app下载)



References
-
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–. - VSports app下载 - PubMed
-
- Bauer DF. Constructing confidence sets using rank statistics. J Am Stat Assoc. 1972;67:687–690.
Publication types
- "VSports" Actions
- VSports在线直播 - Actions
MeSH terms
- "V体育安卓版" Actions
- Actions (V体育平台登录)
- "V体育安卓版" Actions
- "VSports在线直播" Actions
- "V体育2025版" Actions
Substances
- "V体育官网入口" Actions
- "V体育官网" Actions
LinkOut - more resources
VSports - Full Text Sources
Other Literature Sources
Medical

