Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells (VSports在线直播)
- PMID: 19074894
- PMCID: PMC2821873
- DOI: "V体育2025版" 10.1158/0008-5472.CAN-08-0288
VSports注册入口 - Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells
Abstract
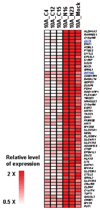
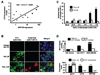
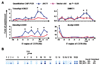
The interplay between histone modifications and promoter hypermethylation provides a causative explanation for epigenetic gene silencing in cancer. Less is known about the upstream initiators that direct this process VSports手机版. Here, we report that the Cystatin M (CST6) tumor suppressor gene is concurrently down-regulated with other loci in breast epithelial cells cocultured with cancer-associated fibroblasts (CAF). Promoter hypermethylation of CST6 is associated with aberrant AKT1 activation in epithelial cells, as well as the disabled INNP4B regulator resulting from the suppression by CAFs. Repressive chromatin, marked by trimethyl-H3K27 and dimethyl-H3K9, and de novo DNA methylation is established at the promoter. The findings suggest that microenvironmental stimuli are triggers in this epigenetic cascade, leading to the long-term silencing of CST6 in breast tumors. Our present findings implicate a causal mechanism defining how tumor stromal fibroblasts support neoplastic progression by manipulating the epigenome of mammary epithelial cells. The result also highlights the importance of direct cell-cell contact between epithelial cells and the surrounding fibroblasts that confer this epigenetic perturbation. Because this two-way interaction is anticipated, the described coculture system can be used to determine the effect of epithelial factors on fibroblasts in future studies. .
Figures



References
-
- Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet. 2006;1:95–101. - PubMed
-
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. - VSports注册入口 - PubMed
-
- Antequera F, Bird A. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol. 1999;9:661–667. - "V体育官网" PubMed
-
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. - V体育官网入口 - PMC - PubMed
-
- Plimack ER, Stewart DJ, Issa JP. Combining epigenetic and cytotoxic therapy in the treatment of solid tumors. J Clin Oncol. 2007;25:4519–4521. - PubMed
Publication types
- VSports app下载 - Actions
- "V体育2025版" Actions
MeSH terms
- "V体育官网" Actions
- "VSports" Actions
- V体育安卓版 - Actions
- "V体育ios版" Actions
- V体育ios版 - Actions
- "VSports在线直播" Actions
- "V体育官网" Actions
- "V体育ios版" Actions
- VSports最新版本 - Actions
- V体育2025版 - Actions
"VSports注册入口" Substances
- V体育官网 - Actions
- VSports app下载 - Actions
Grants and funding
"VSports手机版" LinkOut - more resources
V体育官网 - Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

