A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk
- PMID: 18922928
- PMCID: PMC2672193
- DOI: 10.1158/0008-5472.CAN-08-2129
"V体育安卓版" A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk
VSports手机版 - Abstract
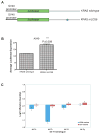
Lung cancer is the leading cause of cancer deaths worldwide, yet few genetic markers of lung cancer risk useful for screening exist. The let-7 family-of-microRNAs (miRNA) are global genetic regulators important in controlling lung cancer oncogene expression by binding to the 3' untranslated regions of their target mRNAs. The purpose of this study was to identify single nucleotide polymorphisms (SNP) that could modify let-7 binding and to assess the effect of such SNPs on target gene regulation and risk for non-small cell lung cancer (NSCLC). let-7 complementary sites (LCS) were sequenced in the KRAS 3' untranslated region from 74 NSCLC cases to identify mutations and SNPs that correlated with NSCLC. The allele frequency of a previously unidentified SNP at LCS6 was characterized in 2,433 people (representing 46 human populations). The frequency of the variant allele is 18. 1% to 20 VSports手机版. 3% in NSCLC patients and 5. 8% in world populations. The association between the SNP and the risk for NSCLC was defined in two independent case-control studies. A case-control study of lung cancer from New Mexico showed a 2. 3-fold increased risk (confidence interval, 1. 1-4. 6; P = 0. 02) for NSCLC cancer in patients who smoked <40 pack-years. This association was validated in a second independent case-control study. Functionally, the variant allele results in KRAS overexpression in vitro. The LCS6 variant allele in a KRAS miRANA complementary site is significantly associated with increased risk for NSCLC among moderate smokers and represents a new paradigm for let-7 miRNAs in lung cancer susceptibility. .
Figures


References
-
- Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–69. - PubMed
-
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics. 2006:38. - PubMed
-
- Saunders MA, Liang H, Wen-Hsiung L. Human polymorphism at microRNAs and microRNA target sites. PNAS. 2007;104:3300–5. - "V体育平台登录" PMC - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed (VSports)
-
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. - PubMed
"VSports在线直播" Publication types
- "V体育ios版" Actions
MeSH terms
- Actions (VSports注册入口)
- "VSports app下载" Actions
- Actions (VSports)
- Actions (V体育官网入口)
- Actions (V体育2025版)
- V体育2025版 - Actions
- "V体育安卓版" Actions
- Actions (VSports最新版本)
- "VSports" Actions
- Actions (V体育官网)
- "VSports最新版本" Actions
- VSports app下载 - Actions
Substances
- Actions (V体育平台登录)
- Actions (V体育2025版)
Grants and funding (VSports手机版)
LinkOut - more resources (VSports手机版)
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

