Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast
- PMID: 18780730
- PMCID: PMC2581965
- DOI: "V体育2025版" 10.1534/genetics.108.092932
VSports app下载 - Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast
Abstract
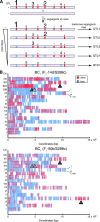
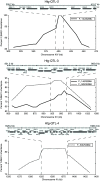
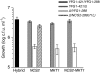
Several quantitative trait loci (QTL) mapping strategies can successfully identify major-effect loci, but often have poor success detecting loci with minor effects, potentially due to the confounding effects of major loci, epistasis, and limited sample sizes. To overcome such difficulties, we used a targeted backcross mapping strategy that genetically eliminated the effect of a previously identified major QTL underlying high-temperature growth (Htg) in yeast. This strategy facilitated the mapping of three novel QTL contributing to Htg of a clinically derived yeast strain. One QTL, which is linked to the previously identified major-effect QTL, was dissected, and NCS2 was identified as the causative gene. The interaction of the NCS2 QTL with the first major-effect QTL was background dependent, revealing a complex QTL architecture spanning these two linked loci VSports手机版. Such complex architecture suggests that more genes than can be predicted are likely to contribute to quantitative traits. The targeted backcrossing approach overcomes the difficulties posed by sample size, genetic linkage, and epistatic effects and facilitates identification of additional alleles with smaller contributions to complex traits. .
VSports注册入口 - Figures




"V体育平台登录" References
-
- Badano, J. L., C. C. Leitch, S. J. Ansley, H. May-Simera, S. Lawson et al., 2006. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature 439 326–330. - PubMed
-
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3 11–21. - PubMed
-
- Botstein, D., and N. Risch, 2003. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat. Genet. 33(Suppl): 228–237. - PubMed
VSports手机版 - Publication types
- Actions (V体育ios版)
MeSH terms
- "V体育2025版" Actions
- "VSports在线直播" Actions
- V体育官网入口 - Actions
- VSports在线直播 - Actions
"VSports在线直播" Substances
- "V体育官网入口" Actions
Associated data
- "VSports手机版" Actions
- "V体育ios版" Actions
- VSports - Actions
- Actions (V体育官网入口)
- Actions
- Actions (V体育平台登录)
- Actions (V体育官网)
- Actions
- Actions (VSports手机版)
- V体育2025版 - Actions
- "V体育官网" Actions
"V体育官网" LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育ios版" Molecular Biology Databases

