"VSports" PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab
- PMID: 18339877
- PMCID: "VSports注册入口" PMC3972216
- DOI: "V体育安卓版" 10.1158/0008-5472.CAN-07-5659
PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab
"VSports最新版本" Erratum in
- Cancer Res. 2008 Aug 15;68(16):6859. Soler, Roman Perez [corrected to Perez-Soler, Roman]
- Cancer Res. 2009 Dec 1;69(23):9156
V体育平台登录 - Abstract
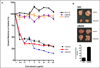
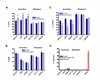
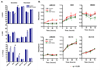
Cetuximab is a monoclonal antibody that targets the human epidermal growth factor receptor (EGFR). Although approved for use in EGFR-overexpressing advanced colorectal cancer, recent studies have shown a lack of association between EGFR overexpression and cetuximab response, requiring the identification of novel biomarkers predictive of response to this agent. To do so, 22 colon cancer cell lines were screened for cetuximab response in vitro and sensitive and resistant lines were identified. In sensitive cell lines, cetuximab induced a G(0)-G(1) arrest without inducing apoptosis VSports手机版. Notably, cetuximab-sensitive but not cetuximab-resistant cell lines were preferentially responsive to EGF-stimulated growth. Whereas neither EGFR protein/mRNA expression nor gene copy number correlated with cetuximab response, examination of the mutation status of signaling components downstream of EGFR showed that cell lines with activating PIK3CA mutations or loss of PTEN expression (PTEN null) were more resistant to cetuximab than PIK3CA wild type (WT)/PTEN-expressing cell lines (14 +/- 5. 0% versus 38. 5 +/- 6. 4% growth inhibition, mean +/- SE; P = 0. 008). Consistently, PIK3CA mutant isogenic HCT116 cells showed increased resistance to cetuximab compared with PIK3CA WT controls. Furthermore, cell lines that were PIK3CA mutant/PTEN null and Ras/BRAF mutant were highly resistant to cetuximab compared with those without dual mutations/PTEN loss (10. 8 +/- 4. 3% versus 38. 8 +/- 5. 9% growth inhibition, respectively; P = 0. 002), indicating that constitutive and simultaneous activation of the Ras and PIK3CA pathways confers maximal resistance to this agent. A priori screening of colon tumors for PTEN expression status and PIK3CA and Ras/BRAF mutation status could help stratify patients likely to benefit from this therapy. .
Figures

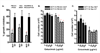
References
-
- Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. - PubMed
-
- Galizia G, Lieto E, Orditura M, et al. Epidermal growth factor receptor (EGFR) expression is associated with a worse prognosis in gastric cancer patients undergoing curative. surgery. World J Surg. 2007;31:1458–1468. - PubMed
-
- McKay JA, Murray LJ, Curran S, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258–2264. - "VSports最新版本" PubMed
-
- Jorissen RN, Walker F, Pouliot N, et al. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. - "VSports app下载" PubMed
-
- Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. - VSports最新版本 - PubMed
"V体育官网入口" MeSH terms
- "V体育安卓版" Actions
- Actions (V体育官网入口)
- Actions (VSports手机版)
- Actions (VSports)
- "VSports" Actions
- Actions (V体育2025版)
- "V体育官网" Actions
- "VSports在线直播" Actions
- Actions (VSports最新版本)
- Actions (VSports最新版本)
- "V体育官网" Actions
- Actions (VSports手机版)
- Actions (VSports最新版本)
Substances
- "V体育平台登录" Actions
- VSports手机版 - Actions
- "V体育平台登录" Actions
- V体育安卓版 - Actions
- Actions (V体育安卓版)
- Actions (VSports在线直播)
Grants and funding
LinkOut - more resources
"V体育2025版" Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

