MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype (VSports注册入口)
- PMID: 17922911
- PMCID: PMC2246288
- DOI: 10.1186/gb-2007-8-10-r214
V体育安卓版 - MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype
Abstract
Background: MicroRNAs (miRNAs), a class of short non-coding RNAs found in many plants and animals, often act post-transcriptionally to inhibit gene expression VSports手机版. .
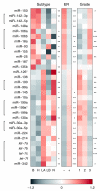
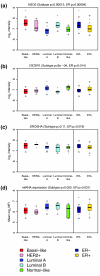
Results: Here we report the analysis of miRNA expression in 93 primary human breast tumors, using a bead-based flow cytometric miRNA expression profiling method V体育安卓版. Of 309 human miRNAs assayed, we identify 133 miRNAs expressed in human breast and breast tumors. We used mRNA expression profiling to classify the breast tumors as luminal A, luminal B, basal-like, HER2+ and normal-like. A number of miRNAs are differentially expressed between these molecular tumor subtypes and individual miRNAs are associated with clinicopathological factors. Furthermore, we find that miRNAs could classify basal versus luminal tumor subtypes in an independent data set. In some cases, changes in miRNA expression correlate with genomic loss or gain; in others, changes in miRNA expression are likely due to changes in primary transcription and or miRNA biogenesis. Finally, the expression of DICER1 and AGO2 is correlated with tumor subtype and may explain some of the changes in miRNA expression observed. .
Conclusion: This study represents the first integrated analysis of miRNA expression, mRNA expression and genomic changes in human breast cancer and may serve as a basis for functional studies of the role of miRNAs in the etiology of breast cancer. Furthermore, we demonstrate that bead-based flow cytometric miRNA expression profiling might be a suitable platform to classify breast cancer into prognostic molecular subtypes. V体育ios版.
Figures




References
-
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. - DOI (VSports) - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. - VSports手机版 - DOI - PubMed
-
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. - "VSports注册入口" DOI - PubMed
Publication types
MeSH terms
- Actions (V体育2025版)
- "V体育安卓版" Actions
- V体育ios版 - Actions
- VSports app下载 - Actions
- V体育官网入口 - Actions
- V体育ios版 - Actions
Substances
- "VSports手机版" Actions
- "VSports手机版" Actions
- "V体育ios版" Actions
LinkOut - more resources
Full Text Sources
"V体育官网入口" Other Literature Sources
VSports手机版 - Medical
Molecular Biology Databases
Research Materials
Miscellaneous

