Optimization of ribosome structure and function by rRNA base modification
- PMID: 17245450
- PMCID: PMC1766470
- DOI: 10.1371/journal.pone.0000174
Optimization of ribosome structure and function by rRNA base modification
Abstract
Background: Translating mRNA sequences into functional proteins is a fundamental process necessary for the viability of organisms throughout all kingdoms of life VSports手机版. The ribosome carries out this process with a delicate balance between speed and accuracy. This work investigates how ribosome structure and function are affected by rRNA base modification. The prevailing view is that rRNA base modifications serve to fine tune ribosome structure and function. .
Methodology/principal findings: To test this hypothesis, yeast strains deficient in rRNA modifications in the ribosomal peptidyltransferase center were monitored for changes in and translational fidelity. These studies revealed allele-specific sensitivity to translational inhibitors, changes in reading frame maintenance, nonsense suppression and aa-tRNA selection. Ribosomes isolated from two mutants with the most pronounced phenotypic changes had increased affinities for aa-tRNA, and surprisingly, increased rates of peptidyltransfer as monitored by the puromycin assay. rRNA chemical analyses of one of these mutants identified structural changes in five specific bases associated with the ribosomal A-site V体育安卓版. .
Conclusions/significance: Together, the data suggest that modification of these bases fine tune the structure of the A-site region of the large subunit so as to assure correct positioning of critical rRNA bases involved in aa-tRNA accommodation into the PTC, of the eEF-1A. aa-tRNA V体育ios版. GTP ternary complex with the GTPase associated center, and of the aa-tRNA in the A-site. These findings represent a direct demonstration in support of the prevailing hypothesis that rRNA modifications serve to optimize rRNA structure for production of accurate and efficient ribosomes. .
Conflict of interest statement
Figures
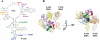


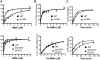
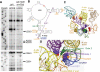
References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, et al. Crystal Structure of the Ribosome at 5.5 A Resolution. Science. 2001;292:883–896. - PubMed
-
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. - PubMed
-
- Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane BG. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. - "VSports最新版本" PubMed
Publication types
"VSports手机版" MeSH terms
- V体育安卓版 - Actions
- "V体育ios版" Actions
- "VSports手机版" Actions
- Actions (V体育ios版)
- Actions (VSports)
- Actions (V体育2025版)
- "VSports最新版本" Actions
- "V体育官网" Actions
- "V体育官网入口" Actions
- V体育官网入口 - Actions
- V体育官网 - Actions
Substances
- "VSports" Actions
- "V体育ios版" Actions
- "V体育官网" Actions
"V体育官网入口" Grants and funding
LinkOut - more resources
Full Text Sources (V体育官网入口)
Molecular Biology Databases
"V体育官网入口" Miscellaneous

