Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture
- PMID: 17093500
- PMCID: PMC1679760 (VSports)
- DOI: 10.1038/sj.emboj.7601412 (VSports注册入口)
Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture
Abstract
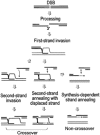
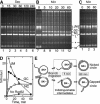
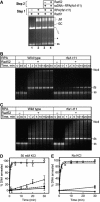
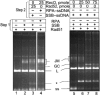
Rad51, Rad52, and RPA play central roles in homologous DNA recombination. Rad51 mediates DNA strand exchange, a key reaction in DNA recombination. Rad52 has two distinct activities: to recruit Rad51 onto single-strand (ss)DNA that is complexed with the ssDNA-binding protein, RPA, and to anneal complementary ssDNA complexed with RPA. Here, we report that Rad52 promotes annealing of the ssDNA strand that is displaced by DNA strand exchange by Rad51 and RPA, to a second ssDNA strand. An RPA that is recombination-deficient (RPA(rfa1-t11)) failed to support annealing, explaining its in vivo phenotype. Escherichia coli RecO and SSB proteins, which are functional homologues of Rad52 and RPA, also facilitated the same reaction, demonstrating its conserved nature. We also demonstrate that the two activities of Rad52, recruiting Rad51 and annealing DNA, are coordinated in DNA strand exchange and second ssDNA capture. VSports手机版.
"V体育平台登录" Figures



References
-
- Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 - "VSports在线直播" PubMed
-
- Carpenter AT (1994) Chiasma function. Cell 77: 957–962 - PubMed
-
- Haber JE, Ira G, Malkova A, Sugawara N (2004) Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos Trans R Soc Lond B Biol Sci 359: 79–86 - PMC (V体育官网) - PubMed
-
- Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A (2004) A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119: 927–940 - PubMed
Publication types (VSports在线直播)
- VSports在线直播 - Actions
MeSH terms
- "VSports" Actions
- Actions (V体育官网入口)
- V体育ios版 - Actions
- Actions (V体育ios版)
Substances
- VSports在线直播 - Actions
- "V体育平台登录" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

