"V体育ios版" The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination
- PMID: 15579681
- PMCID: PMC1448777
- DOI: 10.1534/genetics.103.025700 (V体育平台登录)
The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination (VSports)
Abstract (V体育安卓版)
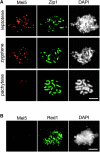
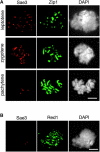
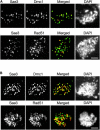
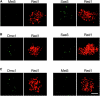
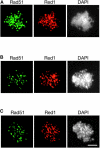
Here we provide evidence that the Mei5 and Sae3 proteins of budding yeast act together with Dmc1, a meiosis-specific, RecA-like recombinase. The mei5 and sae3 mutations reduce sporulation, spore viability, and crossing over to the same extent as dmc1. In all three mutants, these defects are largely suppressed by overproduction of Rad51. In addition, mei5 and sae3, like dmc1, suppress the cell-cycle arrest phenotype of the hop2 mutant VSports手机版. The Mei5, Sae3, and Dmc1 proteins colocalize to foci on meiotic chromosomes, and their localization is mutually dependent. The localization of Rad51 to chromosomes is not affected in either mei5 or sae3. Taken together, these observations suggest that the Mei5 and Sae3 proteins are accessory factors specific to Dmc1. We speculate that Mei5 and Sae3 are necessary for efficient formation of Dmc1-containing nucleoprotein filaments in vivo. .
Figures (V体育官网入口)



V体育2025版 - References
-
- Agarwal, S., and G. S. Roeder, 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. - PubMed
-
- Bishop, D. K., 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092. - PubMed (VSports app下载)
-
- Bishop, D. K., D. Park, L. Xu and N. Kleckner, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. - "VSports手机版" PubMed
Publication types
MeSH terms
- VSports手机版 - Actions
- "V体育ios版" Actions
- "V体育平台登录" Actions
- "V体育平台登录" Actions
- "VSports" Actions
- Actions (V体育平台登录)
- Actions (VSports在线直播)
- "VSports手机版" Actions
Substances
- "V体育平台登录" Actions
- Actions (VSports手机版)
- "VSports注册入口" Actions
- "VSports最新版本" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
"V体育官网入口" Research Materials

