V体育平台登录 - A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain
- PMID: 15310658
- PMCID: PMC515320
- DOI: "V体育官网入口" 10.1101/gr.2743304
A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain
Abstract
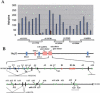
microRNAs (or miRNAs) are small noncoding RNAs (21 to 25 nucleotides) that are processed from longer hairpin RNA precursors and are believed to be involved in a wide range of developmental and cellular processes, by either repressing translation or triggering mRNA degradation (RNA interference). By using a computer-assisted approach, we have identified 46 potential miRNA genes located in the human imprinted 14q32 domain, 40 of which are organized as a large cluster VSports手机版. Although some of these clustered miRNA genes appear to be encoded by a single-copy DNA sequence, most of them are arranged in tandem arrays of closely related sequences. In the mouse, this miRNA gene cluster is conserved at the homologous distal 12 region. In vivo all the miRNAs that we have detected are expressed in the developing embryo (both in the head and in the trunk) and in the placenta, whereas in the adult their expression is mainly restricted to the brain. We also show that the miRNA genes are only expressed from the maternally inherited chromosome and that their imprinted expression is regulated by an intergenic germline-derived differentially methylated region (IG-DMR) located approximately 200 kb upstream from the miRNA cluster. The functions of these miRNAs, which seem only conserved in mammals, are discussed both in terms of epigenetic control and gene regulation during development. .
Figures
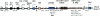

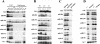

References
-
- Barlow, D.P. 1993. Methylation and imprinting: From host defense to gene regulation? Science 260: 309-310. - PubMed
-
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. - PubMed
-
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. - PubMed
WEB SITE REFERENCES
-
- VSports最新版本 - http://genome.ucsc.edu; Human Genome Project Working Draft.
-
- "V体育平台登录" http://www.tbi.univie.ac.at~/ivo/RNA; RNA fold software.
-
- http://genes.mit.edu/mirscan; software developed by D.P. Bartel and colleagues.
Publication types
- Actions (VSports app下载)
MeSH terms (VSports在线直播)
- "VSports" Actions
- Actions (V体育官网)
- Actions (VSports)
- VSports注册入口 - Actions
- Actions (V体育官网)
- VSports app下载 - Actions
- "V体育2025版" Actions
- Actions (V体育官网)
- "V体育平台登录" Actions
- "V体育官网入口" Actions
- V体育ios版 - Actions
Substances
- V体育官网入口 - Actions
- Actions (VSports手机版)
- VSports - Actions
- "VSports" Actions
- "VSports app下载" Actions
LinkOut - more resources
"VSports在线直播" Full Text Sources
Other Literature Sources
"V体育2025版" Molecular Biology Databases
