V体育平台登录 - GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies
- PMID: 14975175
- PMCID: PMC373441
- DOI: 10.1186/1471-2105-5-16
GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies
Abstract
Background: Microarray and other high-throughput technologies are producing large sets of interesting genes that are difficult to analyze directly VSports手机版. Bioinformatics tools are needed to interpret the functional information in the gene sets. .
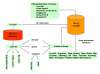
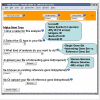
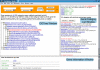
Results: We have created a web-based tool for data analysis and data visualization for sets of genes called GOTree Machine (GOTM). This tool was originally intended to analyze sets of co-regulated genes identified from microarray analysis but is adaptable for use with other gene sets from other high-throughput analyses. GOTree Machine generates a GOTree, a tree-like structure to navigate the Gene Ontology Directed Acyclic Graph for input gene sets. This system provides user friendly data navigation and visualization V体育安卓版. Statistical analysis helps users to identify the most important Gene Ontology categories for the input gene sets and suggests biological areas that warrant further study. GOTree Machine is available online at http://genereg. ornl. gov/gotm/. .
Conclusion: GOTree Machine has a broad application in functional genomic, proteomic and other high-throughput methods that generate large sets of interesting genes; its primary purpose is to help users sort for interesting patterns in gene sets V体育ios版. .
Figures
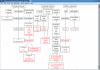
References
-
- Masys DR, Welsh JB, Fink JL, Gribskov M, Klacansky I, Corbeil J. Use of keyword hierarchies to interpret gene expression patterns. Bioinformatics. 2001;17:319–326. doi: 10.1093/bioinformatics/17.4.319. - VSports app下载 - DOI - PubMed
Publication types
- "V体育官网" Actions
"V体育官网入口" MeSH terms
- Actions (VSports app下载)
- V体育平台登录 - Actions
- Actions (VSports注册入口)
- Actions (VSports注册入口)
- V体育2025版 - Actions
- Actions (V体育官网)
- "V体育安卓版" Actions
- V体育平台登录 - Actions
- "V体育平台登录" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
"V体育2025版" Other Literature Sources
Medical

