Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8 (VSports在线直播)
- PMID: 14563679
- PMCID: PMC280615
- DOI: "V体育2025版" 10.1101/gad.1144003
Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8
"V体育2025版" Abstract
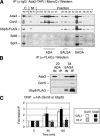
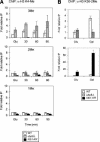
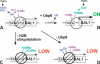
Gene activation and repression regulated by acetylation and deacetylation represent a paradigm for the function of histone modifications. We provide evidence that, in contrast, histone H2B monoubiquitylation and its deubiquitylation are both involved in gene activation. Substitution of the H2B ubiquitylation site at Lys 123 (K123) lowered transcription of certain genes regulated by the acetylation complex SAGA. Gene-associated H2B ubiquitylation was transient, increasing early during activation, and then decreasing coincident with significant RNA accumulation. We show that Ubp8, a component of the SAGA acetylation complex, is required for SAGA-mediated deubiquitylation of histone H2B in vitro. Loss of Ubp8 in vivo increased both gene-associated and overall cellular levels of ubiquitylated H2B. Deletion of Ubp8 lowered transcription of SAGA-regulated genes, and the severity of this defect was exacerbated by codeletion of the Gcn5 acetyltransferase within SAGA VSports手机版. In addition, disruption of either ubiquitylation or Ubp8-mediated deubiquitylation of H2B resulted in altered levels of gene-associated H3 Lys 4 methylation and Lys 36 methylation, which have both been linked to transcription. These results suggest that the histone H2B ubiquitylation state is dynamic during transcription, and that the sequence of histone modifications helps to control transcription. .
Figures (V体育官网)




References
-
- Amerik A.Y., Li, S.-J., and Hochstrasser, M. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381: 981-992. - PubMed
-
- Bhaumik S.R. and Green, M.R. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 15: 1935-1945. - "VSports手机版" PMC - PubMed
-
- Boeger H., Griesenbeck, J., Strattan, J.S., and Kornberg, R.D. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587-1598. - PubMed
Publication types
"V体育2025版" MeSH terms
- V体育2025版 - Actions
- Actions (VSports在线直播)
Substances
- "V体育2025版" Actions
- Actions (V体育平台登录)
VSports在线直播 - Grants and funding
LinkOut - more resources (VSports)
VSports注册入口 - Full Text Sources
Other Literature Sources
V体育官网入口 - Molecular Biology Databases
