The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs (V体育ios版)
- PMID: 14561887
- PMCID: PMC1287059 (VSports app下载)
- DOI: 10.1261/rna.5520403 (V体育2025版)
The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs
Abstract
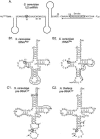
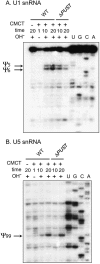
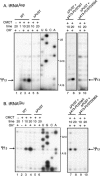
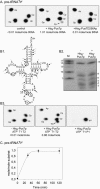
The Saccharomyces cerevisiae Pus7 protein was recently characterized as a novel RNA:pseudouridine (Psi)-synthase acting at position 35 in U2 snRNA. However, U2 snRNA was the only potential substrate tested for this enzyme. In this work, we demonstrated that although Pus7p is responsible for the formation of only one of the six Psi residues present in yeast UsnRNAs, it catalyzes U to Psi conversion at position 13 in cytoplasmic tRNAs and at position 35 in pre-tRNA(Tyr). Sites of RNA modification by Pus7p were identified by analysis of the in vivo RNA modification defects resulting from the absence of active Pus7p production and by in vitro tests using extracts from WT and genetically modified yeast cells. For demonstration of the direct implication of Pus7p in RNA modification, the activity of the WT and mutated Pus7p recombinant proteins was tested on in vitro produced tRNA and pre-tRNA transcripts VSports手机版. Mutation of an aspartic acid residue (D256) that is conserved in all Pus7 homologs abolishes the enzymatic activity both in vivo and in vitro. This suggests the direct involvement of D256 in catalysis. Target sites of Pus7p in RNAs share a common sequence Pu(G/C)UNPsiAPu (Pu = purine, N = any nucleotide), which is expected to be important for substrate recognition. Modification of tRNAs by Pus7p explains the presence of Pus7p homologs in archaea and some bacteria species, which do not have U2 snRNA, and in vertebrates, where Psi34 (equivalent to Psi35 in yeast) formation in U2 snRNA is an H/ACA snoRNA guided process. Our results increase the number of known RNA modification enzymes acting on different types of cellular RNAs. .
"VSports" Figures


References
-
- Adams, A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Ansmant, I. and Motorin, Y. 2001. Identification of RNA modification enzymes using sequence homology. Mol. Biol. (Mosk.) 35: 248–267. - PubMed
-
- Ansmant, I., Massenet, S., Grosjean, H., Motorin, Y., and Branlant, C. 2000. Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of Ψ(2819) in 21S mitochondrial ribosomal RNA. Nucleic Acids Res. 28: 1941–1946. - "V体育2025版" PMC - PubMed
-
- Ansmant, I., Motorin, Y., Massenet, S., Grosjean, H., and Branlant, C. 2001. Identification and characterization of the tRNA: Ψ31-synthase (Pus6p) of Saccharomyces cerevisiae. J. Biol. Chem. 276: 34934–34940. - PubMed
-
- Auxilien, S., Crain, P.F., Trewyn, R.W., and Grosjean, H. 1996. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J. Mol. Biol. 262: 437–458. - V体育2025版 - PubMed
Publication types
- "V体育安卓版" Actions
MeSH terms
- VSports手机版 - Actions
- "VSports在线直播" Actions
- Actions (VSports在线直播)
- VSports在线直播 - Actions
VSports app下载 - Substances
- Actions (VSports在线直播)
- "V体育2025版" Actions
- "VSports手机版" Actions
- Actions (V体育ios版)
- V体育官网 - Actions
- "VSports手机版" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
