A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR (VSports)
- PMID: 12718892
- PMCID: PMC6179153
- DOI: 10.1016/s1097-2765(03)00104-7
A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
"V体育2025版" Abstract
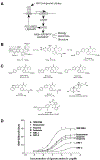
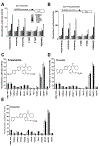
The farnesoid X receptor (FXR) functions as a bile acid (BA) sensor coordinating cholesterol metabolism, lipid homeostasis, and absorption of dietary fats and vitamins. However, BAs are poor reagents for characterizing FXR functions due to multiple receptor independent properties. Accordingly, using combinatorial chemistry we evolved a small molecule agonist termed fexaramine with 100-fold increased affinity relative to natural compounds. Gene-profiling experiments conducted in hepatocytes with FXR-specific fexaramine versus the primary BA chenodeoxycholic acid (CDCA) produced remarkably distinct genomic targets. Highly diffracting cocrystals (1. 78 A) of fexaramine bound to the ligand binding domain of FXR revealed the agonist sequestered in a 726 A(3) hydrophobic cavity and suggest a mechanistic basis for the initial step in the BA signaling pathway. The discovery of fexaramine will allow us to unravel the FXR genetic network from the BA network and selectively manipulate components of the cholesterol pathway that may be useful in treating cholesterol-related human diseases VSports手机版. .
Figures
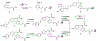



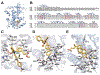
"V体育ios版" Comment in
-
Nuclear receptor ligands and cofactor recruitment: is there a coactivator "on deck"?Mol Cell. 2003 Apr;11(4):850-1. doi: 10.1016/s1097-2765(03)00133-3. Mol Cell. 2003. PMID: 12718871 Review.
References (V体育平台登录)
-
- Blumberg B, and Evans RM (1998). Orphan nuclear receptors—new ligands and new possibilities. Genes Dev. 12, 3149–3155. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. - "V体育ios版" PubMed
-
- Chawla A, Saez E, and Evans RM. (2000). “Don’t know much bile- ology”. Cell. 103, 1–4. - VSports app下载 - PubMed
-
- Chiang JY (2002). Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 23, 443–463. - PubMed
Publication types
MeSH terms
- "V体育官网" Actions
- VSports - Actions
- Actions (VSports app下载)
- VSports在线直播 - Actions
- "V体育官网" Actions
- Actions (VSports在线直播)
- V体育ios版 - Actions
- Actions (V体育官网入口)
- "VSports app下载" Actions
- "V体育2025版" Actions
- "VSports最新版本" Actions
- V体育官网 - Actions
- V体育安卓版 - Actions
- VSports在线直播 - Actions
- "V体育官网入口" Actions
- VSports注册入口 - Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- "V体育安卓版" Actions
Substances
- "VSports注册入口" Actions
- V体育ios版 - Actions
- VSports手机版 - Actions
- V体育2025版 - Actions
V体育官网 - Associated data
- Actions
- "VSports手机版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
V体育2025版 - Other Literature Sources
Molecular Biology Databases

