"VSports手机版" DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair
- PMID: 11940649
- PMCID: PMC133764
- DOI: 10.1128/MCB.22.9.2906-2917.2002
DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair
Abstract
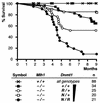
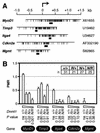
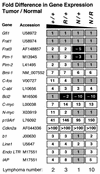
We have introduced DNA methyltransferase 1 (Dnmt1) mutations into a mouse strain deficient for the Mlh1 protein to study the interaction between DNA mismatch repair deficiency and DNA methylation. Mice harboring hypomorphic Dnmt1 mutations showed diminished RNA expression and DNA hypomethylation but developed normally and were tumor free. When crossed to Mlh1(-/-) homozygosity, they were less likely to develop the intestinal cancers that normally arise in this tumor-predisposed, mismatch repair-deficient background. However, these same mice developed invasive T- and B-cell lymphomas earlier and at a much higher frequency than their Dnmt1 wild-type littermates. Thus, the reduction of Dnmt1 activity has significant but opposing outcomes in the development of two different tumor types. DNA hypomethylation and mismatch repair deficiency interact to exacerbate lymphomagenesis, while hypomethylation protects against intestinal tumors. The increased lymphomagenesis in Dnmt1 hypomorphic, Mlh1(-/-) mice may be due to a combination of several mechanisms, including elevated mutation rates, increased expression of proviral sequences or proto-oncogenes, and/or enhanced genomic instability. We show that CpG island hypermethylation occurs in the normal intestinal mucosa, is increased in intestinal tumors in Mlh1(-/-) mice, and is reduced in the normal mucosa and tumors of Dnmt1 mutant mice, consistent with a role for Dnmt1-mediated CpG island hypermethylation in intestinal tumorigenesis VSports手机版. .
Figures


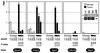

References
-
- Ahuja, N., A. L. Mohan, Q. Li, J. M. Stolker, J. G. Herman, S. R. Hamilton, S. B. Baylin, and J. P. Issa. 1997. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 57:3370-3374. - PubMed
-
- Baker, S. M., C. E. Bronner, L. Zhang, A. W. Plug, M. Robatzek, G. Warren, E. A. Elliott, J. Yu, T. Ashley, N. Arnheim, et al. 1995. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82:309-319. - V体育安卓版 - PubMed
-
- Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. - PubMed
-
- Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168-174. - PubMed
Publication types
MeSH terms
- Actions (V体育官网入口)
- "V体育平台登录" Actions
- "VSports手机版" Actions
- "VSports最新版本" Actions
- V体育ios版 - Actions
- V体育官网入口 - Actions
- "V体育平台登录" Actions
- V体育官网入口 - Actions
- V体育2025版 - Actions
- VSports - Actions
- "VSports" Actions
Substances
- Actions (V体育官网入口)
- VSports app下载 - Actions
- Actions (VSports最新版本)
- "VSports最新版本" Actions
- VSports app下载 - Actions
Grants and funding
LinkOut - more resources
VSports app下载 - Full Text Sources
Other Literature Sources
Molecular Biology Databases
