Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity
- PMID: 10859354
- PMCID: PMC16653
- DOI: VSports - 10.1073/pnas.130192197
"VSports注册入口" Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity
Abstract
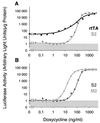
Regulatory elements that control tetracycline resistance in Escherichia coli were previously converted into highly specific transcription regulation systems that function in a wide variety of eukaryotic cells. One tetracycline repressor (TetR) mutant gave rise to rtTA, a tetracycline-controlled transactivator that requires doxycycline (Dox) for binding to tet operators and thus for the activation of P(tet) promoters. Despite the intriguing properties of rtTA, its use was limited, particularly in transgenic animals, because of its relatively inefficient inducibility by doxycycline in some organs, its instability, and its residual affinity to tetO in absence of Dox, leading to elevated background activities of the target promoter. To remove these limitations, we have mutagenized tTA DNA and selected in Saccharomyces cerevisiae for rtTA mutants with reduced basal activity and increased Dox sensitivity. Five new rtTAs were identified, of which two have greatly improved properties. The most promising new transactivator, rtTA2(S)-M2, functions at a 10-fold lower Dox concentration than rtTA, is more stable in eukaryotic cells, and causes no background expression in the absence of Dox VSports手机版. The coding sequences of the new reverse TetR mutants fused to minimal activation domains were optimized for expression in human cells and synthesized. The resulting transactivators allow stringent regulation of target genes over a range of 4 to 5 orders of magnitude in stably transfected HeLa cells. These rtTA versions combine tightness of expression control with a broad regulatory range, as previously shown for the widely applied tTA. .
Figures
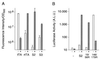
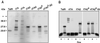
References
-
- Hillen W, Berens C. Annu Rev Microbiol. 1994;48:345–369. - VSports在线直播 - PubMed
-
- Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. - PMC (VSports注册入口) - PubMed
-
- Hinrichs W, Kisker C, Duvel M, Müller A, Tovar K, Hillen W, Saenger W. Science. 1994;264:418–420. - VSports手机版 - PubMed
Publication types
- V体育2025版 - Actions
MeSH terms
- "V体育官网" Actions
- Actions (VSports)
- Actions (VSports app下载)
- "VSports在线直播" Actions
- Actions (VSports)
- V体育安卓版 - Actions
Substances
- "VSports在线直播" Actions
- Actions (VSports注册入口)
- "V体育ios版" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育平台登录" Molecular Biology Databases

