Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins
- PMID: 10330179
- PMCID: PMC104398
- DOI: V体育平台登录 - 10.1128/MCB.19.6.4390
Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins
Abstract
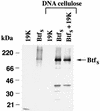
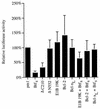
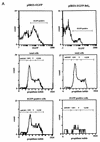
The adenovirus E1B 19,000-molecular-weight (19K) protein is a potent inhibitor of apoptosis and cooperates with E1A to transform primary rodent cells. E1B 19K shows sequence and functional homology to the mammalian antiapoptotic gene product, Bcl-2. Like Bcl-2, the biochemical mechanism of E1B 19K function includes binding to and antagonization of cellular proapoptotic proteins such as Bax, Bak, and Nbk/Bik. In addition, there is evidence that E1B 19K can affect gene expression, but whether this contributes to its antiapoptotic function has not been determined. In an effort to further understand the functions of E1B 19K, we screened for 19K-associated proteins by the yeast two-hybrid system. A novel protein, Btf (Bcl-2-associated transcription factor), that interacts with E1B 19K as well as with the antiapoptotic family members Bcl-2 and Bcl-xL but not with the proapoptotic protein Bax was identified. btf is a widely expressed gene that encodes a protein with homology to the basic zipper (bZip) and Myb DNA binding domains. Btf binds DNA in vitro and represses transcription in reporter assays. E1B 19K, Bcl-2, and Bcl-xL sequester Btf in the cytoplasm and block its transcriptional repression activity. Expression of Btf also inhibited transformation by E1A with either E1B 19K or mutant p53, suggesting a role in either promotion of apoptosis or cell cycle arrest. Indeed, the sustained overexpression of Btf in HeLa cells induced apoptosis, which was inhibited by E1B 19K. Furthermore, the chromosomal localization of btf (6q22-23) maps to a region that is deleted in some cancers, consistent with a role for Btf in tumor suppression. Thus, btf may represent a novel tumor suppressor gene residing in a unique pathway by which the Bcl-2 family can regulate apoptosis. VSports手机版.
Figures




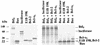


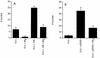

References
-
- Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J-J, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou J-C. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997;277:370–372. - "V体育平台登录" PubMed
-
- Barry M A, Behnke C A, Eastman A. Activation of programmed cell death (apoptosis) by cisplatin, other anticancer drugs, toxins and hyperthermia. Biochem Pharmacol. 1990;40:2353–2362. - PubMed
-
- Bazer L S, Deeg H J. Ultraviolet B-induced DNA fragmentation (apoptosis) in activated T-lymphocytes and Jurkat cells is augmented by inhibition of RNA and protein synthesis. Exp Hematol. 1992;20:80–86. - PubMed
-
- Beg A A, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. - "VSports" PubMed
-
- Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nuñez G, Thompson C. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic death. Cell. 1993;74:597–608. - PubMed
Publication types
VSports app下载 - MeSH terms
- Actions (VSports手机版)
- "V体育安卓版" Actions
- "V体育官网入口" Actions
- V体育官网入口 - Actions
- VSports手机版 - Actions
- "V体育2025版" Actions
- "V体育平台登录" Actions
- V体育ios版 - Actions
- "V体育安卓版" Actions
- Actions (VSports)
- "VSports app下载" Actions
Substances (VSports手机版)
- "VSports注册入口" Actions
- Actions (V体育平台登录)
- "VSports最新版本" Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- VSports手机版 - Actions
- VSports在线直播 - Actions
Associated data
"VSports手机版" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
